3QO4
 
 | | The Crystal Structure of Death Receptor 6 | | Descriptor: | ACETATE ION, SULFATE ION, Tumor necrosis factor receptor superfamily member 21 | | Authors: | Kuester, M, Kemmerzehl, S, Dahms, S.O, Roeser, D, Than, M.E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-18 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
J.Mol.Biol., 409, 2011
|
|
6JJJ
 
 | |
4IXW
 
 | | Halohydrin dehalogenase (HheC) bound to ethyl (2S)-oxiran-2-ylacetate | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase, ethyl (2S)-oxiran-2-ylacetate | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
7OJ1
 
 | | Bacillus subtilis IMPDH in complex with Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Giammarinaro, P.I, Bange, G. | | Deposit date: | 2021-05-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Nat Microbiol, 7, 2022
|
|
4IY1
 
 | | Structure of a 37-fold mutant of halohydrin dehalogenase (HheC) with chloride bound | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
2HYE
 
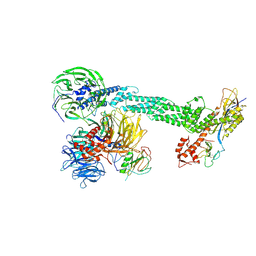 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | Authors: | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | Deposit date: | 2006-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
2ERJ
 
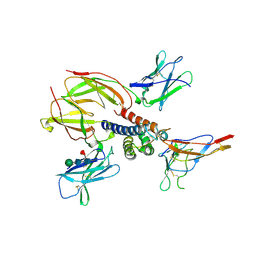 | | Crystal structure of the heterotrimeric interleukin-2 receptor in complex with interleukin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debler, E.W, Stauber, D.J, Wilson, I.A. | | Deposit date: | 2005-10-25 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the IL-2 signaling complex: Paradigm for a heterotrimeric cytokine receptor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4DG9
 
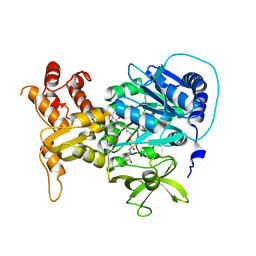 | | Structure of holo-PA1221, an NRPS protein containing adenylation and PCP domains bound to vinylsulfonamide inhibitor | | Descriptor: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, PA1221 | | Authors: | Mitchell, C.A, Shi, C, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of PA1221, a Nonribosomal Peptide Synthetase Containing Adenylation and Peptidyl Carrier Protein Domains.
Biochemistry, 51, 2012
|
|
4IXT
 
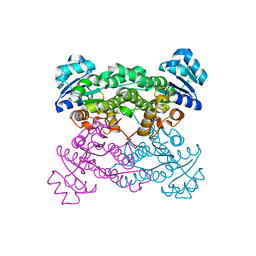 | | Structure of a 37-fold mutant of halohydrin dehalogenase (HheC) bound to ethyl (R)-4-cyano-3-hydroxybutyrate | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase, ethyl (3R)-4-cyano-3-hydroxybutanoate | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
4H0P
 
 | |
3LPW
 
 | | Crystal structure of the FnIII-tandem A77-A78 from the A-band of titin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, A77-A78 domain from Titin | | Authors: | Bucher, R.M, Mayans, O. | | Deposit date: | 2010-02-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the FnIII Tandem A77-A78 points to a periodically conserved architecture in the myosin-binding region of titin
J.Mol.Biol., 401, 2010
|
|
4ZFO
 
 | | J22.9-xi: chimeric mouse/human antibody against human BCMA (CD269) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, J22.9-xi Fab, ... | | Authors: | Marino, S.F, Daumke, O, Olal, D. | | Deposit date: | 2015-04-21 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Potent anti-tumor response by targeting B cell maturation antigen (BCMA) in a mouse model of multiple myeloma.
Mol Oncol, 9, 2015
|
|
3V7Q
 
 | | Crystal structure of B. subtilis YlxQ at 1.55 A resolution | | Descriptor: | CITRIC ACID, POTASSIUM ION, Probable ribosomal protein ylxQ | | Authors: | Baird, N.J, Zhang, J, Hamma, T, Ferre-D'Amare, A.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops.
Rna, 18, 2012
|
|
7P2B
 
 | |
3NX1
 
 | | Crystal structure of Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase | | Descriptor: | Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
5OG1
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
2CGL
 
 | |
6EMI
 
 | | Crystal structure of a variant of human butyrylcholinesterase expressed in bacteria. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cholinesterase, ... | | Authors: | Brazzolotto, X, Igert, A, Guillon, V, Santoni, G, Nachon, F. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development.
Molecules, 22, 2017
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
8CR8
 
 | | human Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, TERBIUM(III) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8D3F
 
 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | Descriptor: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | Authors: | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
8CVZ
 
 | |
8CVY
 
 | |
8CVX
 
 | | Human glycogenin-1 and glycogen synthase-1 complex in the presence of glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Glycogen [starch] synthase, muscle, ... | | Authors: | Liu, Y, Fastman, N.M, Tzitzilonis, C. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural mechanism of human glycogen synthesis by the GYS1-GYG1 complex.
Cell Rep, 40, 2022
|
|
