3VJI
 
 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | Descriptor: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3VJH
 
 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | Descriptor: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3UES
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UET
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UM7
 
 | |
3UTM
 
 | | Crystal structure of a mouse Tankyrase-Axin complex | | Descriptor: | Axin-1, Tankyrase-1 | | Authors: | Cheng, Z, Morrone, S, Xu, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V30
 
 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human RFXANK | | Descriptor: | DNA-binding protein RFX5, DNA-binding protein RFXANK | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3V5E
 
 | | Crystal structure of ClpP from Staphylococcus aureus in the active, extended conformation | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Gersch, M, List, A, Groll, M, Sieber, S. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein.
J.Biol.Chem., 287, 2012
|
|
3V5I
 
 | |
3V9F
 
 | |
3VN9
 
 | | Rifined Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state | | Descriptor: | 9-{5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-beta-L-ribofuranosyl}-9H-purin-6-amine, Dual specificity mitogen-activated protein kinase kinase 6, MAGNESIUM ION | | Authors: | Kinoshita, T, Matsuzaka, H, Nakai, R, Kirii, Y, Yokota, K, Tada, T, Matsumoto, T. | | Deposit date: | 2012-01-05 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state
J.Biochem., 151, 2012
|
|
4DGL
 
 | | Crystal Structure of the CK2 Tetrameric Holoenzyme | | Descriptor: | Casein kinase II subunit alpha, Casein kinase II subunit beta, ZINC ION | | Authors: | Lolli, G, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization.
Acs Chem.Biol., 7, 2012
|
|
2LP0
 
 | |
4AIH
 
 | | Crystal structure of RovA from Yersinia in its free form | | Descriptor: | TRANSCRIPTIONAL REGULATOR SLYA | | Authors: | Quade, N, Mendonca, C, Herbst, K, Heroven, A.K, Heinz, D.W, Dersch, P. | | Deposit date: | 2012-02-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Intrinsic Thermosensing by the Master Virulence Regulator Rova of Yersinia.
J.Biol.Chem., 287, 2012
|
|
4DOR
 
 | |
4DOS
 
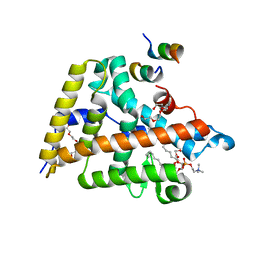 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to DLPC and a Fragment of TIF-2 | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2, ... | | Authors: | Musille, P.M, Ortlund, E.A. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4AK3
 
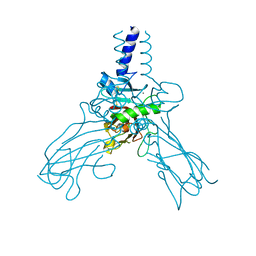 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J.S. | | Deposit date: | 2012-02-21 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AKK
 
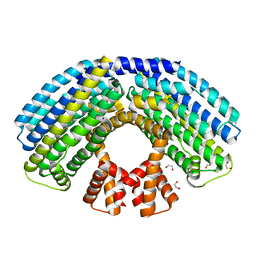 | | Structure of the NasR transcription antiterminator | | Descriptor: | 1,2-ETHANEDIOL, NITRATE REGULATORY PROTEIN | | Authors: | Boudes, M, Lazar, N, Graille, M, Durand, D, Gaidenko, T.A, Stewart, V, van Tilbeurgh, H. | | Deposit date: | 2012-02-24 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | The Structure of the Nasr Transcription Antiterminator Reveals a One-Component System with a Nit Nitrate Receptor Coupled to an Antar RNA-Binding Effector.
Mol.Microbiol., 85, 2012
|
|
4E04
 
 | | RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Bellini, D, Papiz, M.Z. | | Deposit date: | 2012-03-02 | | Release date: | 2012-07-25 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E2J
 
 | | X-Ray Crystal Structure of the Ancestral Glucocorticoid Receptor 2 ligand binding domain in complex with mometasone furoate and TIF-2 coactivator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor 2, FORMIC ACID, GLYCEROL, ... | | Authors: | Kohn, J.A, Deshpande, K, Ortlund, E.A. | | Deposit date: | 2012-03-08 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deciphering Modern Glucocorticoid Cross-pharmacology Using Ancestral Corticosteroid Receptors.
J.Biol.Chem., 287, 2012
|
|
4EFX
 
 | |
4EIW
 
 | | Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Yoshida, M, Morikawa, K. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the whole cytosolic region of ATP-dependent protease FtsH
Mol.Cell, 22, 2006
|
|
4EKW
 
 | | Crystal structure of the NavAb voltage-gated sodium channel (wild-type, 3.2 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein, PHOSPHATE ION | | Authors: | Payandeh, J, Gamal El-Din, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2012-04-10 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of a voltage-gated sodium channel in two potentially inactivated states.
Nature, 486, 2012
|
|
