8RQ1
 
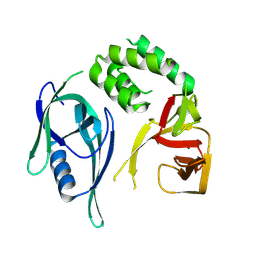 | | Crystal structure of CRBN-midi | | Descriptor: | Protein cereblon, ZINC ION | | Authors: | Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8A5Y
 
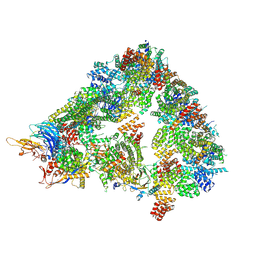 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
3VK1
 
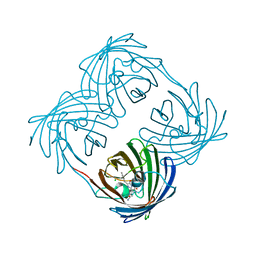 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Wilce, M, Byres, M, Rossjohn, J, Devenish, R.J, Prescott, M. | | Deposit date: | 2011-11-07 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
2VF2
 
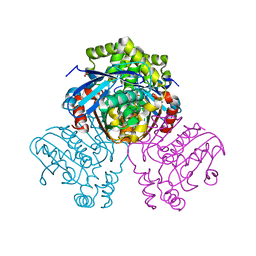 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
8AMZ
 
 | | Spinach 19S proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1 homolog, 26S proteasome non-ATPase regulatory subunit 2 homolog, 26S proteasome regulatory subunit 7, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome
Plant Communications, 3, 2022
|
|
8A61
 
 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
3VNU
 
 | | Complex structure of viral RNA polymerase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
5LAW
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 14 | | Descriptor: | 2-[(3~{S},3'~{a}~{S},6'~{S},6'~{a}~{S})-6-chloranyl-6'-(3-chlorophenyl)-4'-(cyclopropylmethyl)-2-oxidanylidene-spiro[1~{H}-indole-3,5'-3,3~{a},6,6~{a}-tetrahydro-2~{H}-pyrrolo[3,2-b]pyrrole]-1'-yl]ethanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAZ
 
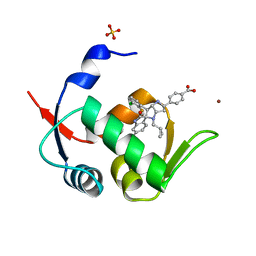 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND BI-0252 | | Descriptor: | 4-[(2~{R},3~{a}~{S},5~{S},6~{S},6~{a}~{S})-6'-chloranyl-6-(3-chloranyl-2-fluoranyl-phenyl)-4-(cyclopropylmethyl)-2'-oxidanylidene-spiro[1,2,3,3~{a},6,6~{a}-hexahydropyrrolo[3,2-b]pyrrole-5,3'-1~{H}-indole]-2-yl]benzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
6PI1
 
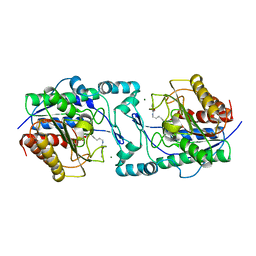 | |
9BR3
 
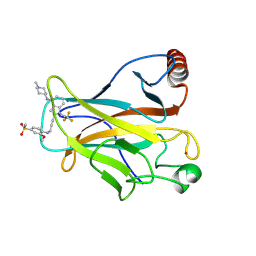 | | Crystal structure of p53 Y220C mutant in complex with PC-10709 | | Descriptor: | 2-{3-[4-(methanesulfonyl)-2-methoxyanilino]prop-1-yn-1-yl}-N-(1-methylpiperidin-4-yl)-1-(2,2,2-trifluoroethyl)-1H-indol-4-amine, Cellular tumor antigen p53, ZINC ION | | Authors: | Abendroth, J, Lorimer, D.D, Vu, B, Tanaka, N. | | Deposit date: | 2024-05-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restoration of the Tumor Suppressor Function of Y220C-Mutant p53 by Rezatapopt, a Small Molecule Reactivator.
Cancer Discov, 2025
|
|
6F9Q
 
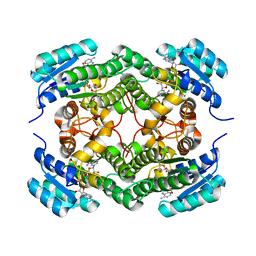 | | Binary complex of a 7S-cis-cis-nepetalactol cyclase from Nepeta mussinii with NAD+ | | Descriptor: | 7S-cis-cis-nepetalactol cyclase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lichman, B.R, Kamileen, M.O, Titchiner, G, Saalbach, G, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
2ITJ
 
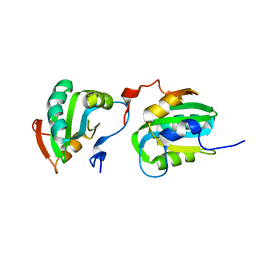 | |
3EI1
 
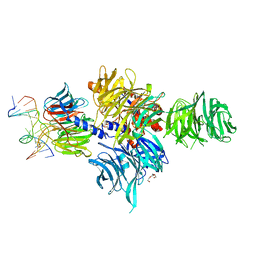 | |
6VHA
 
 | | Singlet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
3V87
 
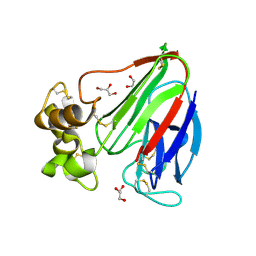 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 1.81 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
9BTP
 
 | | Structure of human SHOC2 in complex with a small molecule inhibitor (R)-5 | | Descriptor: | (2S)-{2-[(4-chloro[1,1'-biphenyl]-3-yl)methoxy]phenyl}[(2-oxo-2,3-dihydro-1,3-benzoxazol-5-yl)amino]acetic acid, Leucine-rich repeat protein SHOC-2, POTASSIUM ION | | Authors: | Dhembi, A, Hauseman, Z.J, King, D. | | Deposit date: | 2024-05-15 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Targeting the SHOC2-RAS interaction in RAS-mutant cancers.
Nature, 2025
|
|
4RV2
 
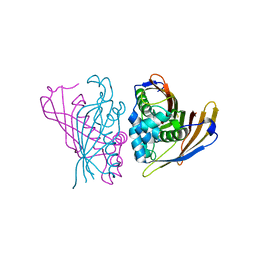 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium smegmatis | | Descriptor: | MaoC family protein, SULFATE ION, UPF0336 protein MSMEG_1340/MSMEI_1302 | | Authors: | Biswas, R, Hazra, D, Dutta, D, Das, A.K. | | Deposit date: | 2014-11-24 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of dehydratase component HadAB complex of mycobacterial FAS-II pathway.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
2J7K
 
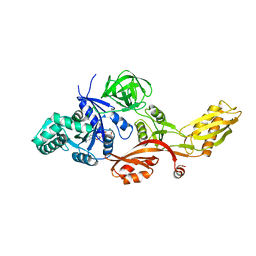 | | Crystal structure of the T84A mutant EF-G:GDPCP complex | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Hansson, S, Logan, D.T. | | Deposit date: | 2006-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New Insights Into the Role of the P-Loop Lysine: Implications from the Crystal Structure of a Mutant EF-G:Gdpcp Complex
To be Published
|
|
6SCM
 
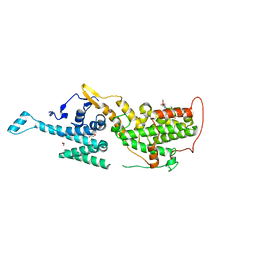 | | SOS1 in Complex with Inhibitor BI-3406 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Son of sevenless homolog 1, ... | | Authors: | Kessler, D, Fischer, G, Ramharter, J. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition.
Cancer Discov, 11, 2021
|
|
1KGT
 
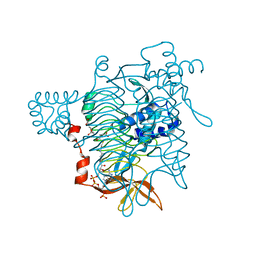 | | Crystal Structure of Tetrahydrodipicolinate N-Succinyltransferase in Complex with Pimelate and Succinyl-CoA | | Descriptor: | 2,3,4,5-TETRAHYDROPYRIDINE-2-CARBOXYLATE N-SUCCINYLTRANSFERASE, PIMELIC ACID, SUCCINYL-COENZYME A | | Authors: | Beaman, T.W, Vogel, K.W, Drueckhammer, D.G, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2001-11-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acyl group specificity at the active site of tetrahydridipicolinate N-succinyltransferase.
Protein Sci., 11, 2002
|
|
3VU1
 
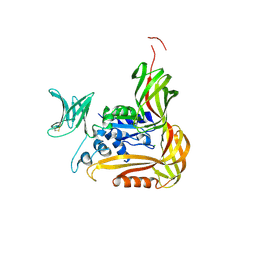 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PhAglB-L, O74088_PYRHO) from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein PH0242 | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
9BTN
 
 | |
1KWT
 
 | | Rat mannose binding protein A (native, MPD) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A | | Authors: | Ng, K.K.S, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
4Z6T
 
 | | Structure of H200N variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-sulfonyl catechol at 1.50 Ang resolution | | Descriptor: | 3,4-dihydroxybenzenesulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
