370D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3'), MAGNESIUM ION | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
369D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
3IWY
 
 | |
4MUR
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUQ
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.364 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1Q74
 
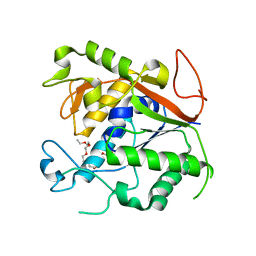 | | The Crystal Structure of 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) | | Descriptor: | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB), 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ZINC ION | | Authors: | Maynes, J.T, Garen, C, Cherney, M.M, Newton, G, Arad, D, Av-Gay, Y, Fahey, R.C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-15 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of 1-D-myo-Inositol 2-Acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) from Mycobacterium tuberculosis Reveals a Zinc Hydrolase with a Lactate Dehydrogenase Fold.
J.Biol.Chem., 278, 2003
|
|
1WVG
 
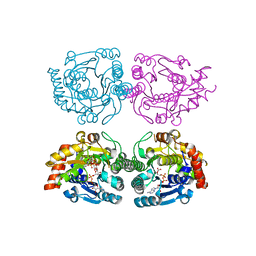 | | Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CDP-glucose 4,6-dehydratase, CYTIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE | | Authors: | Koropatkin, N.M, Holden, H.M. | | Deposit date: | 2004-12-15 | | Release date: | 2005-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi complexed with CDP-D-xylose.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3ZNP
 
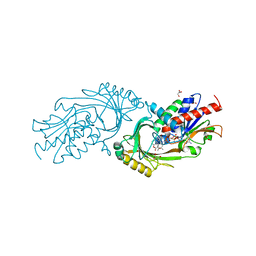 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 3-HYDROXY-2H-CHROMEN-2-ONE, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
3ZNO
 
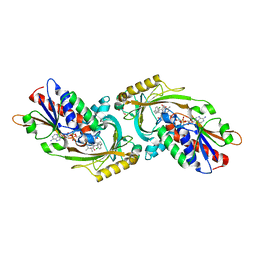 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
3ZNQ
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 3-PHENETHYL-4H-FURO[3,2-B]PYRROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
1HQB
 
 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G, Neuhaus, F.C. | | Deposit date: | 2000-12-14 | | Release date: | 2001-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
4H55
 
 | | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) in complex with beta-d-ribofuranose | | Descriptor: | CALCIUM ION, Concanavalin-Br, D-ALPHA-AMINOBUTYRIC ACID, ... | | Authors: | Salviano, E, Rocha, B.A.M, Cavada, B.S, Delatorre, P, Santi-Gadelha, T, Gadelha, C.A.A, Silva-Filho, J.C, Nobrega, R.B, Farias, D.L. | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) in complex with beta-d-ribofuranose
TO BE PUBLISHED
|
|
1D63
 
 | | CRYSTAL STRUCTURE OF A BERENIL-D(CGCAAATTTGCG) COMPLEX; AN EXAMPLE OF DRUG-DNA RECOGNITION BASED ON SEQUENCE-DEPENDENT STRUCTURAL FEATURES | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Brown, D.G, Sanderson, M.R, Garman, E, Neidle, S. | | Deposit date: | 1992-03-02 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features.
J.Mol.Biol., 226, 1992
|
|
3L35
 
 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
1DK6
 
 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
5C1O
 
 | | Crystal structure of AMP-PNP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4OBZ
 
 | | Structure of Cathepsin D with inhibitor 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide | | Descriptor: | 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3L8G
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with D-glycero-D-manno-heptose 1 ,7-bisphosphate | | Descriptor: | 1,7-di-O-phosphono-L-glycero-beta-D-manno-heptopyranose, D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3NPA
 
 | |
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
372D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
1XX1
 
 | | Structural basis for ion-coordination and the catalytic mechanism of sphingomyelinases D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Murakami, M.T, Tambourgi, D.V, Arni, R.K. | | Deposit date: | 2004-11-03 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases d
J.Biol.Chem., 280, 2005
|
|
1DK9
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
3ZNN
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 4H-THIENO[3,2-B]PYROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
1OPR
 
 | |
