6R8J
 
 | |
6RTJ
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 1 hour of soaking | | Descriptor: | 1-[(dimethylamino)methyl]naphthalen-2-ol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
8WO8
 
 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
7C5O
 
 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
6RGE
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor | | Descriptor: | CITRIC ACID, NAD kinase 1, [(2~{R},3~{R},4~{R},5~{R})-2-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl] dihydrogen phosphate | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor
To Be Published
|
|
8EKX
 
 | | Structure of MBP-Mcl-1 in complex with MIK665 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
1I5V
 
 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
6RMX
 
 | | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Paketuryte, V, Manakova, E, Grazulis, S. | | Deposit date: | 2019-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To Be Published
|
|
5K72
 
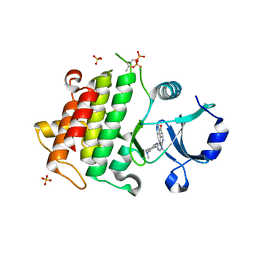 | | IRAK4 in complex with Compound 21 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
8TD0
 
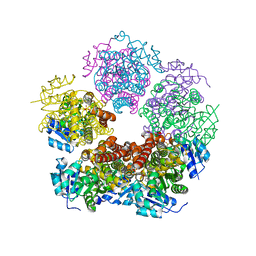 | | Structure of PYCR1 complexed with 5-oxo-7a-phenyl-hexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid | | Descriptor: | (3R,4S,7aR)-5-oxo-7a-phenylhexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
6CZY
 
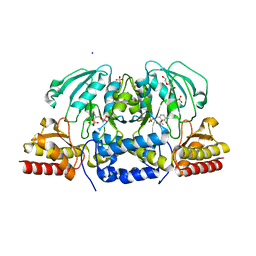 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with Pyridoxamine-5'-phosphate (PMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
8JM0
 
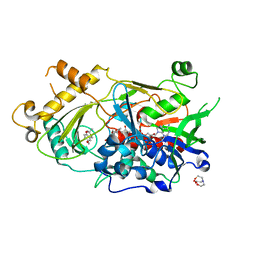 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage | | Descriptor: | (R)-mandelonitrile lyase, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-12 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage
To Be Published
|
|
6UF0
 
 | | Crystal structure of N-(4-((4-methoxy-N-(2,2,2-trifluoroethyl)phenyl)sulfonamido)isoquinolin-1-yl)-N-((4-methoxyphenyl)sulfonyl)glycine bound to human Keap1 Kelch domain | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Kelch-like ECH-associated protein 1, ... | | Authors: | Lazzara, P.R, David, B.P, Ankireddy, A, Richardson, B.G, Dye, K, Ratia, K.M, Reddy, S.P, Moore, T.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Isoquinoline Kelch-like ECH-Associated Protein 1-Nuclear Factor (Erythroid-Derived 2)-like 2 (KEAP1-NRF2) Inhibitors with High Metabolic Stability.
J.Med.Chem., 63, 2020
|
|
5VYS
 
 | | Crystal structure of the WbkC N-formyltransferase (C47S variant) from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
5EHO
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}8-cyclohexyl-~{N}2-[2-methoxy-4-(1-methylpyrazol-4-yl)phenyl]pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach
To Be Published
|
|
3KE3
 
 | |
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
8F9Z
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-14204, an HIV-1 gp120 antagonist | | Descriptor: | (5M)-N-{(1S)-2-amino-1-[5-(hydroxymethyl)-1,3-thiazol-2-yl]ethyl}-5-[5-(trifluoromethyl)pyridin-2-yl]-1H-pyrrole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Antiviral Activity and Crystal Structures of HIV-1 gp120 Antagonists.
Int J Mol Sci, 23, 2022
|
|
8FA0
 
 | |
6ZPU
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain with inserted symmetry molecule C-terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Angiotensin-converting enzyme open for business: structural insights into the subdomain dynamics.
Febs J., 288, 2021
|
|
6LL1
 
 | | Oxygen-exposed reduced terminal oxygenase in carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxygen-exposed reduced terminal oxygenase in carbazole 1,9a-dioxygenase
To Be Published
|
|
6CJY
 
 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 5-[(7H-purin-6-yl)amino]-1H-isoindol-1-one, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CK3
 
 | | Co-crytsal Structure of MNK2 in Complex With an Inhibitor | | Descriptor: | (3R)-3-methyl-5-[(pyrimidin-4-yl)amino]-2,3-dihydro-1H-isoindol-1-one, CHLORIDE ION, MAP kinase-interacting serine/threonine-protein kinase 2, ... | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CW7
 
 | | E. coli DHFR product complex with (6S)-5,6,7,8-TETRAHYDROFOLATE | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-30 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6CZX
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP internal aldimine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
