6QFX
 
 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 10) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
9F3A
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-325 | | Descriptor: | 3C-like proteinase nsp5, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
8SG2
 
 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
9FHQ
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR04 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-6-oxidanylidene-pyrimidin-5-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9FJX
 
 | |
9CDY
 
 | | Crystal structure of DLK with inhibitor bound | | Descriptor: | 1,2-ETHANEDIOL, 3-{(3M)-3-[6-amino-5-(trifluoromethoxy)pyridin-3-yl]-1-ethyl-7-oxo-1,7-dihydro-6H-pyrazolo[3,4-c]pyridin-6-yl}-1,5-anhydro-3,4-dideoxy-2-O-methyl-D-erythro-pentitol, MAGNESIUM ION, ... | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2024-06-25 | | Release date: | 2025-02-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In Silico Enabled Discovery of KAI-11101, a Preclinical DLK Inhibitor for the Treatment of Neurodegenerative Disease and Neuronal Injury.
J.Med.Chem., 68, 2025
|
|
7T8V
 
 | | Co-crystal structure of Chaetomium glucosidase I with EB-0159 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
9EZ6
 
 | | Complex of a mutant of the SARS-CoV-2 main protease Mpro with the nsp14/15 substrate peptide. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Battistutta, R, Fornasier, E, Giachin, G. | | Deposit date: | 2024-04-10 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Allostery in homodimeric SARS-CoV-2 main protease.
Commun Biol, 7, 2024
|
|
7TAN
 
 | | Structure of VRK1 C-terminal tail bound to nucleosome core particle | | Descriptor: | Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, Histone H3.2, ... | | Authors: | Spangler, C.J, Budziszewski, G.R, McGinty, R.K. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity.
Nucleic Acids Res., 50, 2022
|
|
7T6W
 
 | | Crystal structure of Chaetomium Glucosidase I (apo) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, GLYCEROL, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
9EY9
 
 | | Yeast 20S proteasome in complex with a sybactin derivative (PheSyr) | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Praeve, L, Kuttenlochner, W, Tabak, W.W.A, Langer, C, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2024-04-09 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bioengineering of syrbactin megasynthetases for immunoproteasome inhibitor production
Chem, 10, 2024
|
|
8YIC
 
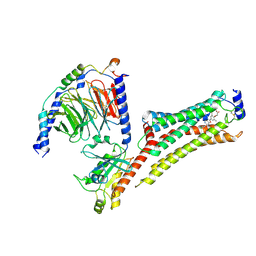 | | SAR247799-bound S1PR1-Gi protein complex | | Descriptor: | Endolysin,Sphingosine 1-phosphate receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Yu, J.J, Shao, Z.H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | SAR247799-bound S1PR1 in complex with Gi heterotrimer
To Be Published
|
|
8C3W
 
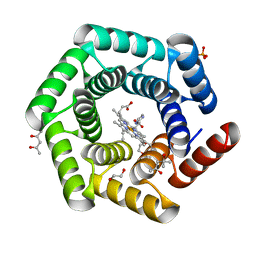 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
9EZ4
 
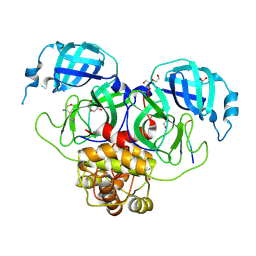 | | Complex of a mutant of the SARS-CoV-2 main protease Mpro with the nsp5/6 substrate peptide. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTAMINE, ... | | Authors: | Battistutta, R, Fornasier, E, Giachin, G. | | Deposit date: | 2024-04-10 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allostery in homodimeric SARS-CoV-2 main protease.
Commun Biol, 7, 2024
|
|
9AVA
 
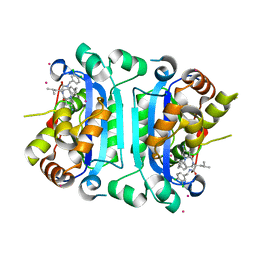 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9F2V
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR02 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{R},3~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
5LMP
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA translation pre-initiation complex (state-1C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.35 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
8HCD
 
 | | Crystal structure of mTREX1-DNA product complex (dNMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
7TCS
 
 | | M379A mutant tyrosine phenol-lyase complexed with L-methionine | | Descriptor: | (4Z)-4-({[(1E)-1-carboxy-3-(methylsulfanyl)propylidene]azaniumyl}methylidene)-2-methyl-5-[(phosphonooxy)methyl]-1,4-dihydropyridin-3-olate, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | M379A Mutant Tyrosine Phenol-lyase from Citrobacter freundii Has Altered Conformational Dynamics.
Chembiochem, 23, 2022
|
|
8HCH
 
 | | Crystal structure of mTREX1-Uridine complex | | Descriptor: | MAGNESIUM ION, Three-prime repair exonuclease 1, URIDINE | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
7SQO
 
 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
9AXC
 
 | | Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
5LKK
 
 | |
9AYL
 
 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
