2PQD
 
 | | A100G CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
8BIP
 
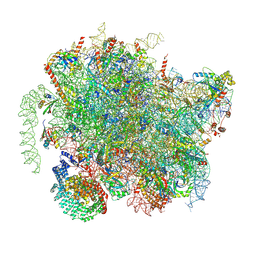 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
4CAS
 
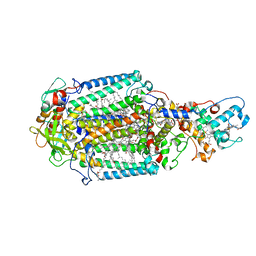 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
2PR8
 
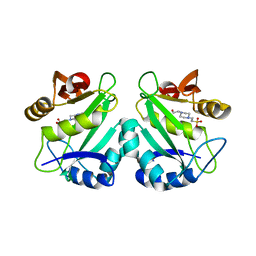 | | crystal structure of aminoglycoside N-acetyltransferase AAC(6')-Ib11 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside 6-N-acetyltransferase type Ib11 | | Authors: | Maurice, F, Broutin, I, Podglajen, I, Benas, P, Collatz, E, Dardel, F. | | Deposit date: | 2007-05-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance.
Embo Rep., 9, 2008
|
|
7XLY
 
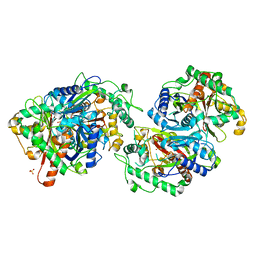 | | Crystal structure of FadA2 (Rv0243) from the fatty acid metabolic pathway of Mycobacterium tuberculosis | | Descriptor: | Probable acetyl-CoA acyltransferase FadA2 (3-ketoacyl-CoA thiolase) (Beta-ketothiolase), SULFATE ION | | Authors: | Singh, R, Kundu, P, Singh, B.K, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of FadA2 thiolase from Mycobacterium tuberculosis and prediction of its substrate specificity and membrane-anchoring properties.
Febs J., 290, 2023
|
|
5KJV
 
 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5D0N
 
 | | Crystal structure of maize PDRP bound with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyruvate, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-03 | | Release date: | 2016-02-24 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
5CUP
 
 | |
8BJQ
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
2PSW
 
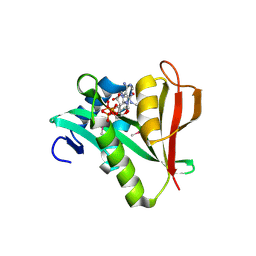 | | Human MAK3 homolog in complex with CoA | | Descriptor: | COENZYME A, N-acetyltransferase 13 | | Authors: | Walker, J.R, Schuetz, A, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human MAK3 homolog.
To be Published
|
|
2HG3
 
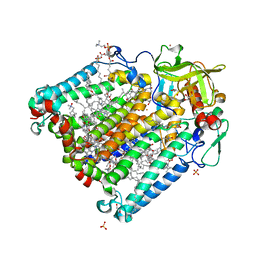 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with brominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HG9
 
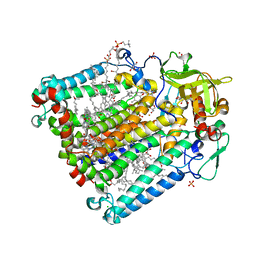 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with tetrabrominated phosphatidylcholine | | Descriptor: | (7R,18S,19R)-18,19-DIBROMO-7-{[(9S,10S)-9,10-DIBROMOOCTADECANOYL]OXY}-4-HYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-P HOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2Q6I
 
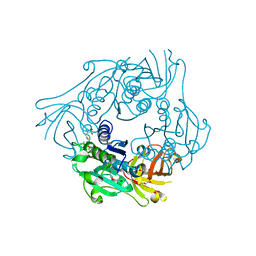 | | salL with ClDA and LMet | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, DI(HYDROXYETHYL)ETHER, Hypothetical protein, ... | | Authors: | Noel, J.P, Pojer, F. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
3LZC
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
4EWG
 
 | |
2HRO
 
 | | Structure of the full-lenght Enzyme I of the PTS system from Staphylococcus carnosus | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase, SULFATE ION | | Authors: | Marquez, J.A, Reinelt, S, Koch, B, Engelman, R, Hengstenberg, W, Scheffzek, K. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the full-length enzyme I of the phosphoenolpyruvate-dependent sugar phosphotransferase system
J.Biol.Chem., 281, 2006
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
4F0Y
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-IG from Acinetobacter haemolyticus, apo | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
2HH1
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
5KTM
 
 | |
5KTS
 
 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound citraconate and Fe4S4 cluster | | Descriptor: | (~{Z})-2-methylbut-2-enedioic acid, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
2Q4V
 
 | | Ensemble refinement of the protein crystal structure of thialysine n-acetyltransferase (SSAT2) from Homo sapiens | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 2 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
5ZMW
 
 | | Crystal structure of the E309Q mutant of SR Ca2+-ATPase in E2(TG) | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Ogawa, H, Hirata, A, Tsueda, J, Toyoshima, C. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of the E2 to E1 transition in Ca2+pump revealed by crystal structures of gating residue mutants.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZHQ
 
 | |
4F32
 
 | | Crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase II from Burkholderia vietnamiensis in complex with platencin | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-23 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
