2MS3
 
 | |
2MS4
 
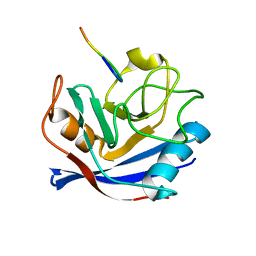 | | Cyclophilin a complexed with a fragment of crk-ii | | Descriptor: | Peptide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Jankowski, W, Saleh, T, Rossi, P, Kalodimos, C. | | Deposit date: | 2014-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway.
Nat.Chem.Biol., 12, 2016
|
|
2MS5
 
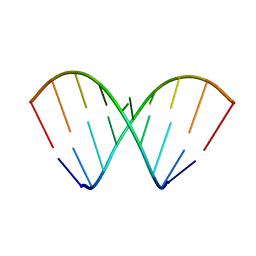 | | Structural dynamics of double-helical RNA having CAG motif | | Descriptor: | RNA_(5'-R(P*CP*CP*GP*CP*AP*GP*CP*GP*G)-3') | | Authors: | Kumar, A, Tawani, A. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights Reveal the Dynamics of the Repeating r(CAG) Transcript Found in Huntington's Disease (HD) and Spinocerebellar Ataxias (SCAs)
Plos One, 10, 2015
|
|
2MS6
 
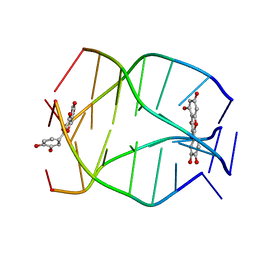 | | Human Telomeric G-quadruplex DNA sequence (TTAGGGT)4 complexed with Flavonoid Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DNA_(5'-D(*TP*TP*AP*GP*GP*GP*T)-3') | | Authors: | Kumar, A, Tawani, A. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the interaction of Flavonoids with Human Telomeric Sequence
Sci Rep, 5, 2015
|
|
2MS7
 
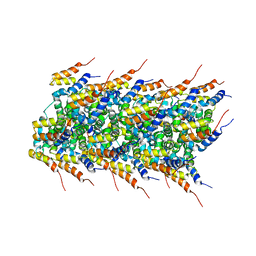 | | High-resolution solid-state NMR structure of the helical signal transduction filament MAVS CARD | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | He, L, Bardiaux, B, Spehr, J, Luehrs, T, Ritter, C. | | Deposit date: | 2014-07-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of helical filaments by solid-state NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2MS8
 
 | | Solution NMR structure of MAVS CARD | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Spehr, J, He, L, Luehrs, T, Ritter, C. | | Deposit date: | 2014-07-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure determination of helical filaments by solid-state NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2MS9
 
 | | Solution structure of a G-quadruplex | | Descriptor: | DNA (28-MER) | | Authors: | Chung, W.J, Heddi, B, Schmitt, E, Lim, K.W, Mechulam, Y, Phan, A.T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a left-handed DNA G-quadruplex
Proc.Natl.Acad.Sci.USA, 2015
|
|
2MSA
 
 | |
2MSB
 
 | | STRUCTURE OF A C-TYPE MANNOSE-BINDING PROTEIN COMPLEXED WITH AN OLIGOSACCHARIDE | | Descriptor: | CALCIUM ION, MANNOSE-BINDING PROTEIN-A, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Weis, W.I, Drickamer, K, Hendrickson, W.A. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a C-type mannose-binding protein complexed with an oligosaccharide.
Nature, 360, 1992
|
|
2MSC
 
 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSD
 
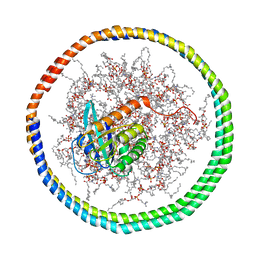 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSF
 
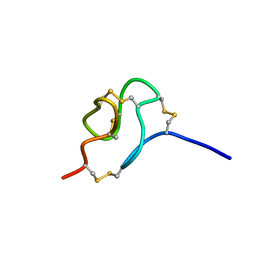 | | NMR SOLUTION STRUCTURE OF SCORPION VENOM TOXIN Ts11 (TsPep1) FROM Tityus serrulatus | | Descriptor: | Peptide TsPep1 | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Cremonez, C.M, Peigneur, S, Cassoli, J.S, Dutra, A.A.A, Waelkens, E, Pimenta, A.M.C, De Lima, M.H, Tytgat, J, Arantes, E.C. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Elucidation of Peptide Ts11 Shows Evidence of a Novel Subfamily of Scorpion Venom Toxins.
Toxins (Basel), 8, 2016
|
|
2MSG
 
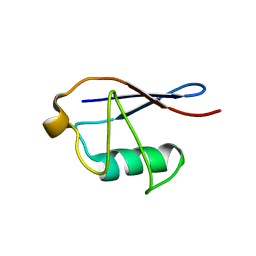 | | Solid-state NMR structure of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Lakomek, N, Habenstein, B, Loquet, A, Shi, C, Giller, K, Wolff, S, Becker, S, Fasshuber, H, Lange, A. | | Deposit date: | 2014-08-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural heterogeneity in microcrystalline ubiquitin studied by solid-state NMR.
Protein Sci., 24, 2015
|
|
2MSI
 
 | |
2MSJ
 
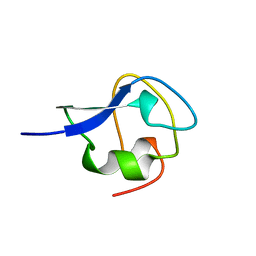 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N46S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
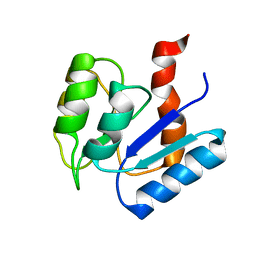
2MSK
 
 | |
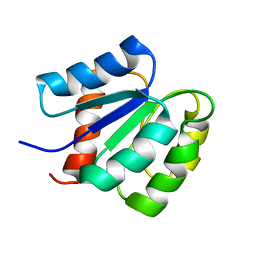
2MSL
 
 | |
2MSN
 
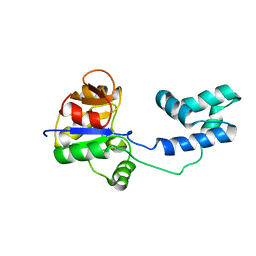 | | NMR structure of a putative phosphoglycolate phosphatase (NP_346487.1) from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MSO
 
 | | Solution study of cGm9a | | Descriptor: | Conotoxin Gm9.1 | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2MSP
 
 | | MAJOR SPERM PROTEIN, BETA ISOFORM, ENGINEERED C59S/T90C MUTANT, PUTATIVE SUBFILAMENT STRUCTURE, PH 8.5 | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Bullock, T.L, Mccoy, A.J, Stewart, M. | | Deposit date: | 1997-12-19 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for amoeboid motility in nematode sperm.
Nat.Struct.Biol., 5, 1998
|
|
2MSQ
 
 | | Solution study of cBru9a | | Descriptor: | Conotoxin cBru9a | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2MSR
 
 | | Solution structure of LEDGF/p75 IBD in complex with MLL1 peptide (140-160) | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein | | Authors: | Cermakova, K, Tesina, P, Demeulemeester, J, El Ashkar, S, Mereau, H, Schwaller, J, Rezacova, P, Veverka, V, De Rijck, J. | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Validation and Structural Characterization of the LEDGF/p75-MLL Interface as a New Target for the Treatment of MLL-Dependent Leukemia.
Cancer Res., 74, 2014
|
|
2MSS
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2MST
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
