7OLI
 
 | | Crystal structure of Pab-AGOG in complex with 8-oxoguanosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-8-OXOGUANOSINE, N-glycosylase/DNA lyase | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
8CN2
 
 | | Pa.FabF-C164Q in complex with N-isobutyl-1H-pyrazole-3-carboxamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, ~{N}-(2-methylpropyl)-1~{H}-pyrazole-3-carboxamide | | Authors: | Georgiou, C, Brenk, R. | | Deposit date: | 2023-02-21 | | Release date: | 2024-01-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Towards new antibiotics: P. aeruginosa FabF ligands discovered by crystallographic fragment screening followed by hit expansion.
Eur.J.Med.Chem., 291, 2025
|
|
8CAW
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with agrocin84 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, [(2R,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.256 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
9G0K
 
 | | CysG(N-16) in complex with SAH (metal free, cocrystallization in presence of EDTA) from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-07-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
7OKV
 
 | | Crystal structure of soluble EPCR after exposure to the nonionic surfactant Polysorbate 20 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Erausquin, E, Dichiara, M.G, Lopez-Sagaseta, J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of a broad lipid repertoire associated to the endothelial cell protein C receptor (EPCR).
Sci Rep, 12, 2022
|
|
6NFH
 
 | | BTK in complex with inhibitor 8-(2,3-dihydro-1H-inden-5-yl)-2-({4-[(2S)-3-(dimethylamino)-2-hydroxypropoxy]phenyl}amino)-5,8-dihydropteridine-6,7-dione | | Descriptor: | 1,2-ETHANEDIOL, 8-(2,3-dihydro-1H-inden-5-yl)-2-({4-[(2S)-3-(dimethylamino)-2-hydroxypropoxy]phenyl}amino)-5,8-dihydropteridine-6,7-dione, Tyrosine-protein kinase BTK | | Authors: | Mirzadegan, T, Damm-Ganamet, K.L. | | Deposit date: | 2018-12-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Accelerating Lead Identification by High Throughput Virtual Screening: Prospective Case Studies from the Pharmaceutical Industry.
J.Chem.Inf.Model., 59, 2019
|
|
7ANT
 
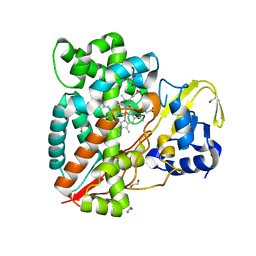 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7AO7
 
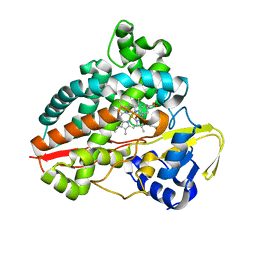 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
6NE9
 
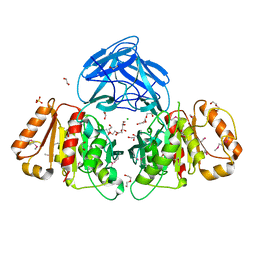 | | Bacteroides intestinalis acetyl xylan esterase (BACINT_01039) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Pereira, G.V, Cann, I. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Degradation of complex arabinoxylans by human colonic Bacteroidetes
Nat Commun, 2021
|
|
7XDH
 
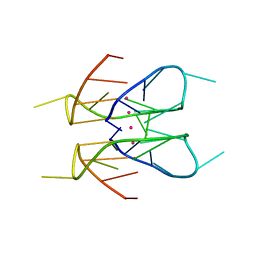 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | Descriptor: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | Authors: | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
7SUA
 
 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
8ZKD
 
 | | The Crystal Structure of the RON from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Macrophage-stimulating protein receptor beta chain, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the RON from Biortus.
To Be Published
|
|
5IHV
 
 | |
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
7QYP
 
 | | The structure of T. forsythia NanH | | Descriptor: | 1,2-ETHANEDIOL, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
8EN6
 
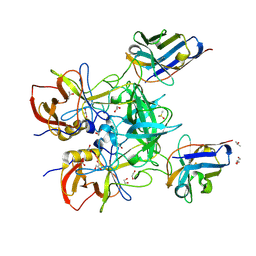 | | Structure of GII.4 norovirus in complex with Nanobody 76 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GII.4 P domain, ... | | Authors: | Kher, G, Sabin, C, Pancera, M, Koromyslova, A, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
7P35
 
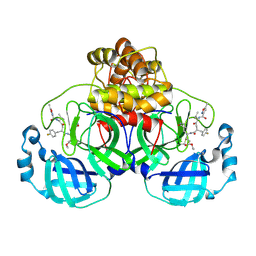 | | Structure of the SARS-CoV-2 3CL protease in complex with rupintrivir | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Fabrega-Ferrer, M, Perez-Saavedra, J, Herrera-Morande, A, Coll, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
8VTH
 
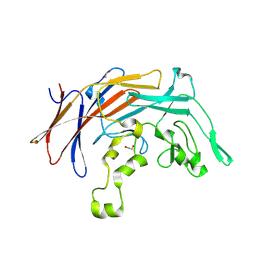 | | DUF2169 domain-containing protein from Vibrio xiamenensis | | Descriptor: | 1,2-ETHANEDIOL, DUF2169 domain-containing protein | | Authors: | Sachar, K, Whitney, J.C, Prehna, G, Kim, Y. | | Deposit date: | 2024-01-26 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved chaperone protein is required for the formation of a noncanonical type VI secretion system spike tip complex.
J.Biol.Chem., 301, 2025
|
|
8CQV
 
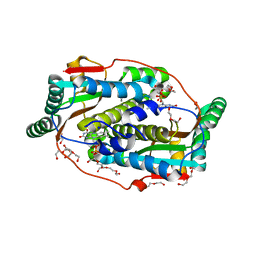 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CND
 
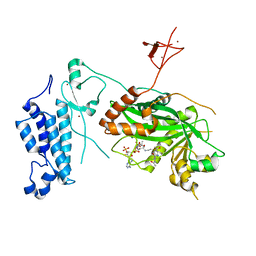 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
6W1X
 
 | | Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
1F28
 
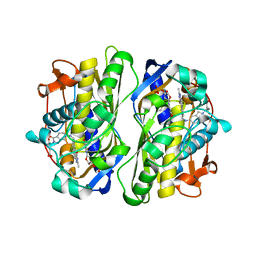 | | CRYSTAL STRUCTURE OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII BOUND TO DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Anderson, A.C, O'Neil, R.H, Surti, T.S, Stroud, R.M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking.
Chem.Biol., 8, 2001
|
|
6WAX
 
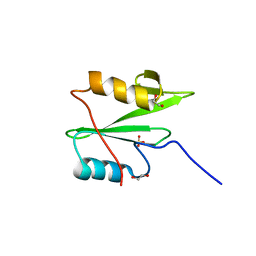 | | C-terminal SH2 domain of p120RasGAP | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein 1, SULFATE ION | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
8WD2
 
 | | The Crystal Structure of p53 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Lu, Y. | | Deposit date: | 2023-09-14 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of p53 from Biortus.
To Be Published
|
|
