8CIS
 
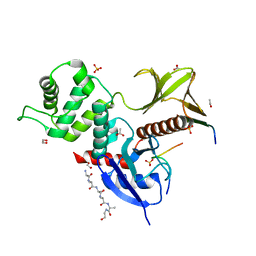 | | The FERM domain of human moesin with two bound peptides identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C3P, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
6MTS
 
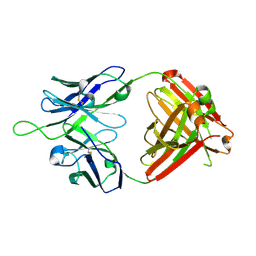 | | Crystal structure of VRC43.03 Fab | | Descriptor: | Antibody VRC43.03 Fab heavy chain, Antibody VRC43.03 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
8H3H
 
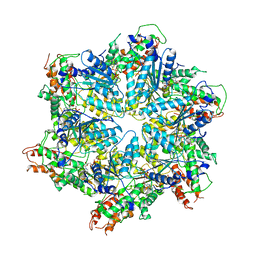 | | Human ATAD2 Walker B mutant, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
6MK4
 
 | |
8JHW
 
 | |
7O33
 
 | | Crystal structure of the anti-PAS Fab 3.1 in complex with its epitope peptide | | Descriptor: | APSA epitope peptide, anti-PAS Fab 3.1 chimeric heavy chain, anti-PAS Fab 3.1 chimeric light chain | | Authors: | Schilz, J, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
8E8L
 
 | | 9H2 Fab-poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
1E7U
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
7OE4
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-methyl-4-propionyl-1H-pyrrole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Bromodomain-containing protein 2, ... | | Authors: | Chung, C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Fragment-based Scaffold Hopping: Identification of Potent, Selective, and Highly Soluble Bromo and Extra Terminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
8T90
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-(3OH)IP5 | | Descriptor: | (1R,2R,3R,4S,5R,6R)-6-hydroxycyclohexane-1,2,3,4,5-pentayl pentakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
6NAB
 
 | |
7QCS
 
 | | PALS1/MPP5 PDZ domain in complex with SARS-CoV-2_E PBM peptide | | Descriptor: | Envelope small membrane protein, Protein PALS1 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
6MJM
 
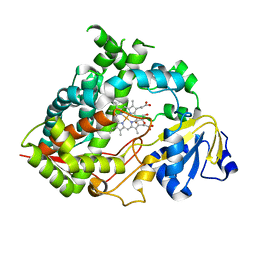 | | Substrate Free Cytochrome P450 3A5 (CYP3A5) | | Descriptor: | Cytochrome P450 3A5, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2018-09-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site differences between substrate-free and ritonavir-bound cytochrome P450 (CYP) 3A5 reveal plasticity differences between CYP3A5 and CYP3A4.
J.Biol.Chem., 294, 2019
|
|
6N2C
 
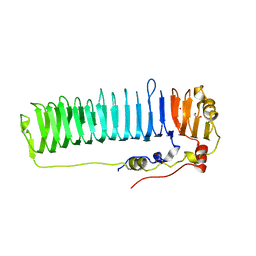 | |
6N3L
 
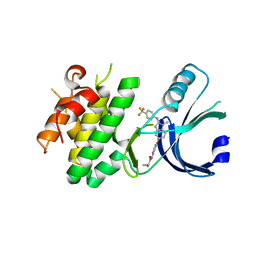 | | Identification of novel, potent and selective GCN2 inhibitors as first-in-class anti-tumor agents | | Descriptor: | N-{6-[(1-methyl-2-{[4-(trifluoromethyl)phenyl]amino}-1H-benzimidazol-5-yl)oxy]pyrimidin-4-yl}cyclopropanecarboxamide, eIF-2-alpha kinase GCN2,eIF-2-alpha kinase GCN2 | | Authors: | Hoffman, I.D, Fujimoto, J, Kurasawa, O, Takagi, T, Klein, M.G, Kefala, G, Ding, S.C, Cary, D.R, Mizojiri, R. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of Novel, Potent, and Orally Available GCN2 Inhibitors with Type I Half Binding Mode.
Acs Med.Chem.Lett., 10, 2019
|
|
7QRZ
 
 | | Crystal structure of B30.2 PRYSPRY domain of MID2 | | Descriptor: | 1,2-ETHANEDIOL, Probable E3 ubiquitin-protein ligase MID2, SULFATE ION | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural analysis of TRIM family PRYSPRY domains and its implications for E3-ligand design.
J Struct Biol X, 12, 2025
|
|
7QS2
 
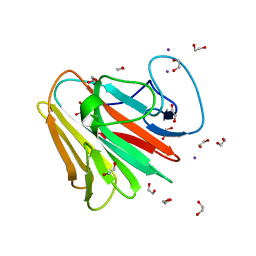 | | Crystal structure of B30.2 PRYSPRY domain of TRIM15 | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, TRIM15 | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of TRIM family PRYSPRY domains and its implications for E3-ligand design.
J Struct Biol X, 12, 2025
|
|
7QS3
 
 | | Crystal structure of B30.2 PRYSPRY domain of TRIM16 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tripartite motif-containing protein 16 | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of TRIM family PRYSPRY domains and its implications for E3-ligand design.
J Struct Biol X, 12, 2025
|
|
8HE6
 
 | | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution | | Descriptor: | All3014 protein, CALCIUM ION, MAGNESIUM ION | | Authors: | Chatterjee, A, Singh, P.K, Singh, T.P, Marina, A, Sharma, S, Rai, L.C. | | Deposit date: | 2022-11-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution
To Be Published
|
|
7OO0
 
 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8EDY
 
 | |
6VLM
 
 | |
6VJB
 
 | |
6VJJ
 
 | | Crystal Structure of wild-type KRAS4b (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
