5C1A
 
 | | p97-N750D/R753D/M757D/Q760D in complex with ATP-gamma-S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
5CJ2
 
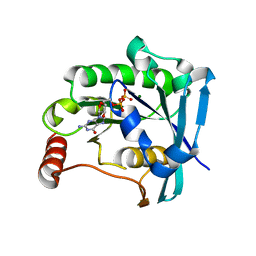 | | Ran GDP Y39A mutant triclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
6CZ5
 
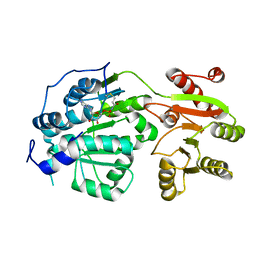 | | Crystal structure of small molecule AMP-acrylamide covalently bound to DDX3 S228C | | Descriptor: | 5'-O-[(R)-hydroxy(propanoylamino)phosphoryl]adenosine, ATP-dependent RNA helicase DDX3X | | Authors: | Barkovich, K.J, Moore, M.K, Hu, Q, Shokat, K.M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemical genetic inhibition of DEAD-box proteins using covalent complementarity.
Nucleic Acids Res., 46, 2018
|
|
5CLQ
 
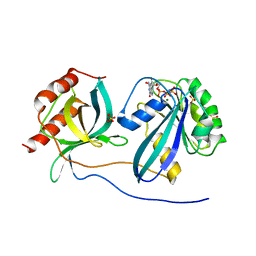 | | Ran Y39A in complex with GPPNHP and RanBD1 | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
6CKS
 
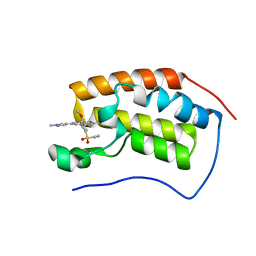 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | 4-[5-(ethylsulfonyl)-2-methoxyphenyl]-2-methyl-6-(1-methyl-1H-pyrazol-4-yl)isoquinolin-1(2H)-one, Bromodomain-containing protein 4 | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6CM2
 
 | | SAMHD1 HD domain bound to decitabine triphosphate | | Descriptor: | 6-amino-3-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-3,4-dihydro-1,3,5-triazin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Oellerich, T, Schneider, C, Thomas, D, Knecht, K.M, Buzovetsky, O, Kaderali, L, Schliemann, C, Bohnenberger, H, Angenendt, L, Hartmann, W, Wardelmann, E, Rothenburger, T, Mohr, S, Scheich, S, Comoglio, F, Wilke, A, Strobel, P, Serve, H, Michaelis, M, Ferreiros, N, Geisslinger, G, Xiong, Y, Keppler, O.T, Cinatl, J. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Selective inactivation of hypomethylating agents by SAMHD1 provides a rationale for therapeutic stratification in AML.
Nat Commun, 10, 2019
|
|
5C56
 
 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
6CJ1
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG071 | | Descriptor: | 1,2-ETHANEDIOL, 11-[(2R)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, 11-[(2S)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CSW
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor | | Descriptor: | (7R)-5-butyl-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7,8-dimethyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Chiodi, C.G, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor
To Be Published
|
|
6CWA
 
 | |
6CMM
 
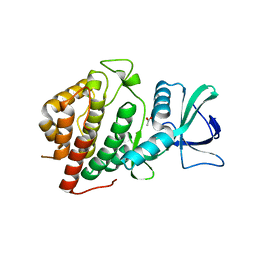 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor
To Be Published
|
|
6CZV
 
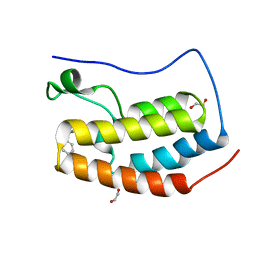 | | BRD4(BD1) complexed with 2759 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-5-(3,5-dimethyl-1,2-oxazol-4-yl)pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
7W3D
 
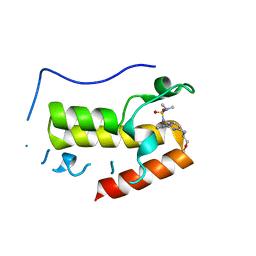 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
5CLL
 
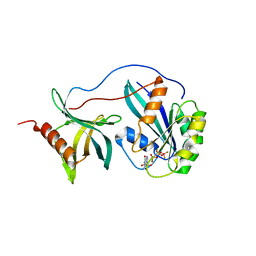 | | Truncated Ran wild type in complex with GDP-BeF and RanBD1 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CFW
 
 | |
7W3L
 
 | | Crystal structure of LSD1 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
6D9X
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | 2-phenyl-6,7-dihydro-5H-pyrrolo[1,2-a]imidazole, SODIUM ION, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
6DD4
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-dipropyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, Santiago, A.S, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropyl-dihydropteridine inhibitor
To Be Published
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WL4
 
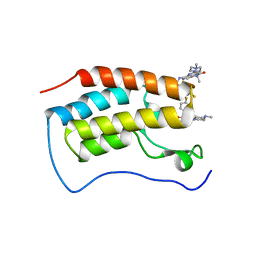 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor SLP-50 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-ethyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]-2,4-bis(fluoranyl)benzenesulfonamide | | Authors: | Zhang, C, Wang, C, Li, W, Zhang, Y, Xu, Y, Sun, L. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis, and anticancer evaluation of ammosamide B with pyrroloquinoline derivatives as novel BRD4 inhibitors.
Bioorg.Chem., 127, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
6DHS
 
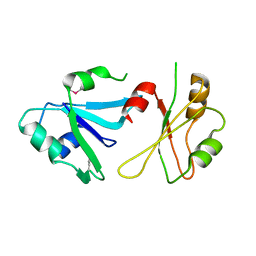 | | Structure of hnRNP H qRRM1,2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Conformational Dynamics Encoded by the Inter-qRRM linker of hnRNP H.
J. Am. Chem. Soc., 2018
|
|
5CIW
 
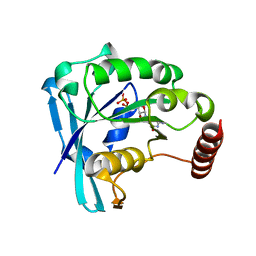 | | Ran GDP Y39A mutant monoclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CIQ
 
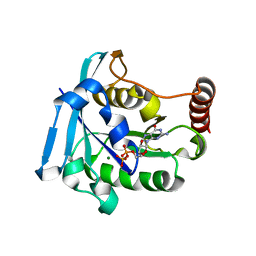 | | Ran GDP wild type tetragonal crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
