1A6P
 
 | |
1A7Q
 
 | | FV FRAGMENT OF MOUSE MONOCLONAL ANTIBODY D1.3 (BALB/C, IGG1, K) HIGH AFFINITY EXPRESSED VARIANT CONTAINING SER26L->GLY, ILE29L->THR, GLU81L->ASP, THR97L->SER, PRO240H->LEU, ASP258H->ALA, LYS281H->GLU, ASN283H->ASP AND LEU312H->VAL | | Descriptor: | IGG1-KAPPA D1.3 FV (HEAVY CHAIN), IGG1-KAPPA D1.3 FV (LIGHT CHAIN) | | Authors: | Marks, C, Henrick, K, Winter, G. | | Deposit date: | 1998-03-16 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structures of D1.3 Fv Mutants
To be Published
|
|
1AH1
 
 | | CTLA-4, NMR, 20 STRUCTURES | | Descriptor: | CTLA-4, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Metzler, W.J, Bajorath, J, Fenderson, W, Shaw, S.-Y, Peach, R, Constantine, K.L, Naemura, J, Leytze, G, Lavoie, T.B, Mueller, L, Linsley, P.S. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28.
Nat.Struct.Biol., 4, 1997
|
|
1A7O
 
 | |
1A6U
 
 | | B1-8 FV FRAGMENT | | Descriptor: | B1-8 FV (HEAVY CHAIN), B1-8 FV (LIGHT CHAIN) | | Authors: | Simon, T, Henrick, K, Hirshberg, M, Winter, G. | | Deposit date: | 1998-03-03 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Fv Fragment and its (4-Hydroxy-3-Nitrophenyl)Acetate Complex of Murine B1-8 Antibody
To be Published
|
|
2LVE
 
 | | RECOMBINANT LEN | | Descriptor: | RLEN | | Authors: | Schiffer, M, Pokkuluri, P.R. | | Deposit date: | 1998-05-13 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A domain flip as a result of a single amino-acid substitution.
Structure, 6, 1998
|
|
2MMX
 
 | |
2MCP
 
 | |
2N7B
 
 | | Solution structure of the human Siglec-8 lectin domain in complex with 6'sulfo sialyl Lewisx | | Descriptor: | 3-aminopropan-1-ol, N-acetyl-alpha-neuraminic acid-(2-3)-6-O-sulfo-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2OCW
 
 | |
2OR7
 
 | | Tim-2 | | Descriptor: | ACETATE ION, T-cell immunoglobulin and mucin domain-containing protein 2 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
2OR8
 
 | | Tim-1 | | Descriptor: | ACETATE ION, Hepatitis A virus cellular receptor 1 homolog | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
2P1Y
 
 | |
2OTU
 
 | | Crystal structure of Fv polyglutamine complex | | Descriptor: | Fv heavy chain variable domain, Fv light chain variable domain, peptide antigen | | Authors: | Li, P. | | Deposit date: | 2007-02-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Implications of the structure of a poly-Gln/anti-poly-Gln complex for disease progression and therapy
To be Published
|
|
2OYP
 
 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
2NMS
 
 | |
2N4I
 
 | | The solution structure of Skint-1, a critical determinant of dendritic epidermal gamma-delta T cell selection | | Descriptor: | Selection and upkeep of intraepithelial T-cells protein 1 | | Authors: | Salim, M, Knowles, T.J, Hart, R, Mohammed, F, Woodward, M.J, Willcox, C.R, Overduin, M, Hayday, A.C, Willcox, B.E. | | Deposit date: | 2015-06-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Putative Receptor Binding Surface on Skint-1, a Critical Determinant of Dendritic Epidermal T Cell Selection.
J.Biol.Chem., 291, 2016
|
|
2Q20
 
 | |
2M2D
 
 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
2MKW
 
 | |
2MCG
 
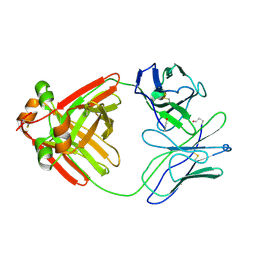 | |
2Q1E
 
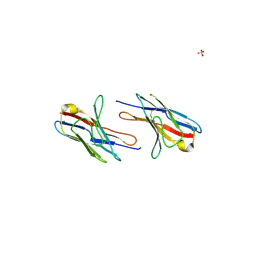 | |
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2N7A
 
 | | Solution structure of the human Siglec-8 lectin domain | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2Q87
 
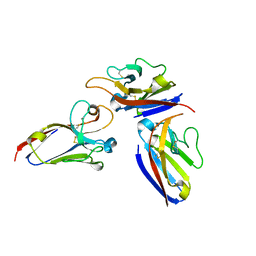 | |
