6MH1
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH HU-10, A 1,4,5-Trisubstituted Imidazole Analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-(3,5-dimethylphenyl)-4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]pyrimidin-2-amine | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2018-09-17 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for the N-Terminal Bromodomain-and-Extra-Terminal-Family Selectivity of a Dual Kinase-Bromodomain Inhibitor.
J.Med.Chem., 61, 2018
|
|
8D8L
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 3) | | Descriptor: | 15S ribosomal RNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Burnside, C, Harper, N, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
2JTN
 
 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
6SXR
 
 | | E221Q mutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
3IA7
 
 | | Crystal Structure of CalG4, the Calicheamicin Glycosyltransferase | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalG4 | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5HMX
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 10 | | Descriptor: | 2,2'-(5-(thiophen-2-yl)-1,3-phenylene)diacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
5CTY
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2C0H
 
 | | X-ray structure of beta-mannanase from blue mussel Mytilus edulis | | Descriptor: | MANNAN ENDO-1,4-BETA-MANNOSIDASE, SULFATE ION | | Authors: | Larsson, A.M, Anderson, L, Xu, B, Munoz, I.G, Uson, I, Janson, J.-C, Stalbrand, H, Stahlberg, J. | | Deposit date: | 2005-09-02 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-Dimensional Crystal Structure and Enzymic Characterization of Beta-Mannanase Man5A from Blue Mussel Mytilus Edulis.
J.Mol.Biol., 357, 2006
|
|
2BNX
 
 | | Crystal structure of the dimeric regulatory domain of mouse diaphaneous-related formin (DRF), mDia1 | | Descriptor: | CHLORIDE ION, DIAPHANOUS PROTEIN HOMOLOG 1 | | Authors: | Otomo, T, Otomo, C, Tomchick, D.R, Machius, M, Rosen, M.K. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Rho Gtpase-Mediated Activation of the Formin Mdia1
Mol.Cell, 18, 2005
|
|
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
7W96
 
 | |
6J2Z
 
 | | AtFKBP53 N-terminal Nucleoplasmin Domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-03 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
7VVH
 
 | |
3GB2
 
 | | GSK3beta inhibitor complex | | Descriptor: | 2-methyl-5-(3-{4-[(S)-methylsulfinyl]phenyl}-1-benzofuran-5-yl)-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D. | | Deposit date: | 2009-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-{3-[4-(Alkylsulfinyl)phenyl]-1-benzofuran-5-yl}-5-methyl-1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta with good brain permeability.
J.Med.Chem., 52, 2009
|
|
1BWI
 
 | | THE 1.8 A STRUCTURE OF MICROBATCH OIL DROP GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6UYZ
 
 | | Crystal structure of K46-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
5CVN
 
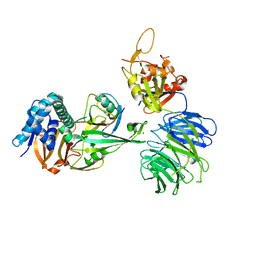 | | WDR48 (2-580):USP46~ubiquitin ternary complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, WD repeat-containing protein 48, ... | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5HJ5
 
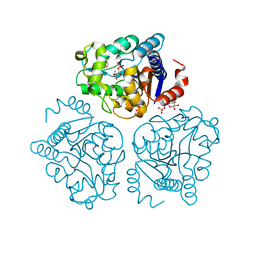 | | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE -6-PHOSPHATE
To Be Published
|
|
2N4D
 
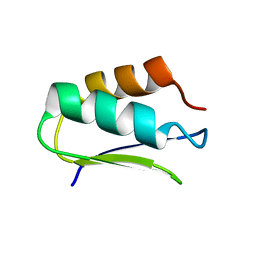 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
5OJZ
 
 | |
2JXF
 
 | | The solution structure of HCV NS4B(40-69) | | Descriptor: | Genome polyprotein | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2007-11-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Determinant for Membrane Association in Hepatitis C Virus Nonstructural Protein 4B
J.Virol., 83, 2009
|
|
2N0S
 
 | | HADDOCK model of ferredoxin and [FeFe] hydrogenase complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Fe-hydrogenase, Ferredoxin, ... | | Authors: | Rumpel, S, Siebel, J, Fares, C, Reijerse, E, Lubitz, W. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Complex of Ferredoxin and [FeFe] Hydrogenase from Chlamydomonas reinhardtii.
Chembiochem, 16, 2015
|
|
8HY8
 
 | | Bacterial STING from Epilithonimonas lactis | | Descriptor: | CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8D8K
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 2) | | Descriptor: | 15S ribosomal RNA, 37S ribosomal protein MRP1, mitochondrial, ... | | Authors: | Burnside, C, Harper, N.J, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
3FRO
 
 | | Crystal structure of Pyrococcus abyssi glycogen synthase with open and closed conformations | | Descriptor: | 1,5-anhydro-D-fructose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GlgA glycogen synthase, ... | | Authors: | Diaz, A, Guinovart, J.J, Fita, I, Ferrer, J.C. | | Deposit date: | 2009-01-08 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lyase activity of glycogen synthase: Is an elimination/addition mechanism a possible reaction pathway for retaining glycosyltransferases?
IUBMB Life, 64, 2012
|
|
