4BPQ
 
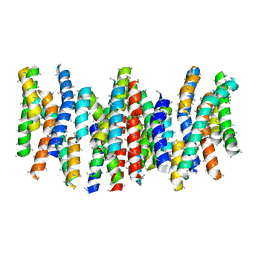 | | Structure and substrate induced conformational changes of the secondary citrate-sodium symporter CitS revealed by electron crystallography | | Descriptor: | CITRATE:SODIUM SYMPORTER | | Authors: | Kebbel, F, Kurz, M, Arheit, M, Gruetter, M.G, Stahlberg, H. | | Deposit date: | 2013-05-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Structure and Substrate-Induced Conformational Changes of the Secondary Citrate/Sodium Symporter Cits Revealed by Electron Crystallography.
Structure, 21, 2013
|
|
2PQ3
 
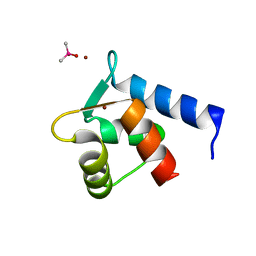 | | N-Terminal Calmodulin Zn-Trapped Intermediate | | Descriptor: | CACODYLATE ION, Calmodulin, ZINC ION | | Authors: | Warren, J.T, Guo, Q, Tang, W.J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A 1.3-A structure of zinc-bound N-terminal domain of calmodulin elucidates potential early ion-binding step.
J.Mol.Biol., 374, 2007
|
|
2PQC
 
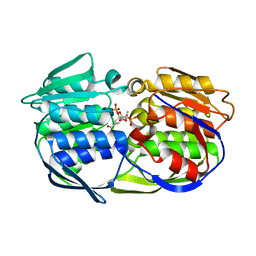 | | CP4 EPSPS liganded with (R)-phosphonate tetrahedral reaction intermediate analog | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
4BTY
 
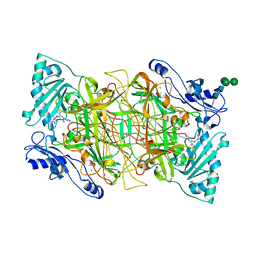 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-(4-methylpiperazin-1-yl)phenylamino]-2-(4-chlorophenyl)-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, ... | | Authors: | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
2PR6
 
 | |
4EV4
 
 | | Crystal structure of serratia fonticola carbapenemase SFC-1 E166A mutant with the acylenzyme intermediate of meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1 | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
4EMS
 
 | |
2OZA
 
 | | Structure of p38alpha complex | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | White, A, Pargellis, C.A, Studts, J.M, Werneburg, B.G, Farmer II, B.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of MAPK-activated protein kinase 2:p38 assembly
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PD1
 
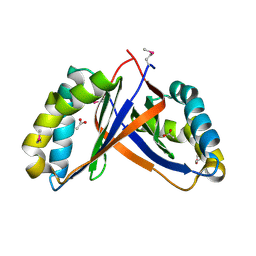 | | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea | | Descriptor: | ACETATE ION, Hypothetical protein | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea.
To be Published
|
|
4EDO
 
 | |
4EEQ
 
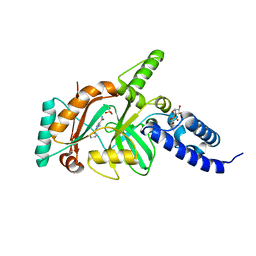 | | Crystal structure of E. faecalis DNA ligase with inhibitor | | Descriptor: | 4-amino-2-(cyclopentyloxy)pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wang, T, Charifson, P, Wei, Y. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis and Activity Evaluation of Potent NAD+ DNA Ligase Inhibitors as Potential Antibacterial Agents.
To be Published
|
|
2PJS
 
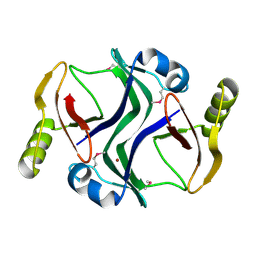 | | Crystal structure of Atu1953, protein of unknown function | | Descriptor: | Uncharacterized protein Atu1953, ZINC ION | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily protein Atu1953, protein of unknown function
To be Published
|
|
2H0U
 
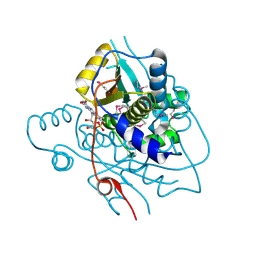 | | Crystal structure of NAD(P)H-flavin oxidoreductase from Helicobacter pylori | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-flavin oxidoreductase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-15 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NAD(P)H-flavin oxidoreductase from Helicobacter pylori
To be Published
|
|
2H3G
 
 | |
2PQB
 
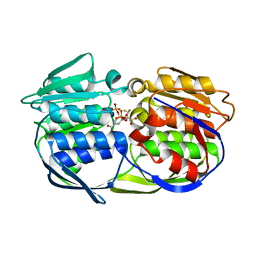 | | CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2GVS
 
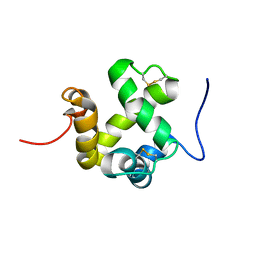 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
2H9M
 
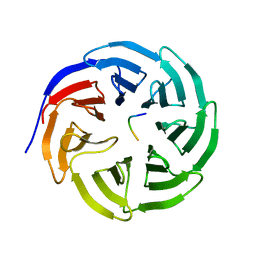 | | WDR5 in complex with unmodified H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2HA2
 
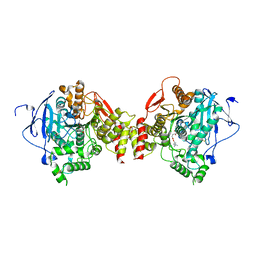 | | Crystal structure of mouse acetylcholinesterase complexed with succinylcholine | | Descriptor: | 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
2H26
 
 | | human CD1b in complex with endogenous phosphatidylcholine and spacer | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Vallina, A.T, Guillet, V, Mourey, L. | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b.
Embo J., 25, 2006
|
|
4DUI
 
 | | DARPIN D1 binding to tubulin beta chain (not in complex) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1 | | Authors: | Pecqueur, L, Duellberg, C, Dreier, B, Wang, Q, Jiang, C, Pluckthun, A, Surrey, T, Gigant, B, Knossow, M. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | An Anti-Tubulin Darpin Caps the Microtubule Plus-End
To be Published
|
|
4DUN
 
 | | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, Putative phenazine biosynthesis PhzC/PhzF protein, ... | | Authors: | Brunzelle, J.S, Wawrzak, W, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630)
To be Published
|
|
2HEG
 
 | | Phospho-Aspartyl Intermediate Analogue of Apha class B acid phosphatase/phosphotransferase | | Descriptor: | Class B acid phosphatase, MAGNESIUM ION | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-06-21 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
4DX0
 
 | | Structure of the 14-3-3/PMA2 complex stabilized by a pyrazole derivative | | Descriptor: | 14-3-3-like protein E, 4-[(4R)-4-(4-nitrophenyl)-6-oxidanylidene-3-phenyl-1,4-dihydropyrrolo[3,4-c]pyrazol-5-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Richter, A, Rose, R, Hedberg, C, Waldmann, H, Ottmann, C. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Optimised Small-Molecule Stabiliser of the 14-3-3-PMA2 Protein-Protein Interaction.
Chemistry, 18, 2012
|
|
4E0N
 
 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form II) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DEZ
 
 | | Structure of MsDpo4 | | Descriptor: | DNA polymerase IV 1 | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2012-01-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The PAD region in the mycobacterial DinB homologue MsPolIV exhibits positional heterogeneity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
