6SOD
 
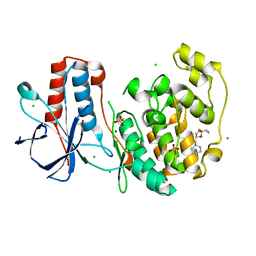 | | Fragment N14056a in complex with MAP kinase p38-alpha | | Descriptor: | 1-[[(3~{S})-1,4-dioxaspiro[4.5]decan-3-yl]methyl]piperidine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
7A4X
 
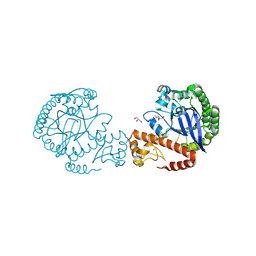 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with dimethyl sulfoxide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nguyen, D, Nguyen, T.X.P, Heine, A, Klebe, G. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
3V52
 
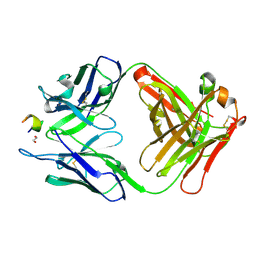 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, ... | | Authors: | Mage, M.G, Dolan, M.A, Wang, R, Boyd, L.F, Revilleza, M.J, Robinson, H, Natarajan, K, Myers, N.B, Hansen, T.H, Margulies, D.H. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
6SR4
 
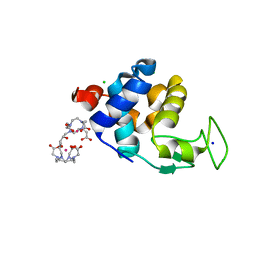 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 112 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
9G7B
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and loreclezole | | Descriptor: | 1-[(~{Z})-2-chloranyl-2-(2,4-dichlorophenyl)ethenyl]-1,2,4-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-07-20 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | CryoEM structure of human rho1 GABAA receptor in complex with neurosteroid
To be published
|
|
7A3G
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, 91 | | Descriptor: | 1-[3-(7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-6-ylmethyl)phenyl]-3,3-diethyl-azetidine-2,4-dione, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
6DLU
 
 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
3MWQ
 
 | |
1E5J
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH METHYL-4II-S-ALPHA-CELLOBIOSYL-4II-THIO-BETA-CELLOBIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Fort, S, Varrot, A, Schulein, M, Cottaz, S, Driguez, H, Davies, G.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mixed-Linkage Cellooligosaccharides: A New Class of Glycoside Hydrolase Inhibitors
Chembiochem, 2, 2001
|
|
5FD5
 
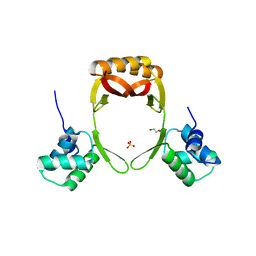 | | manganese uptake regulator | | Descriptor: | 1,2-ETHANEDIOL, Ferric uptake regulation protein, SULFATE ION | | Authors: | Bellini, D, Lebedev, A, Keegan, R, Walsh, M.A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of an apo metal-free manganese uptake regulator, mur
To Be Published
|
|
6DO6
 
 | | NMR solution structure of wild type apo hFABP1 at 308 K | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
3EJG
 
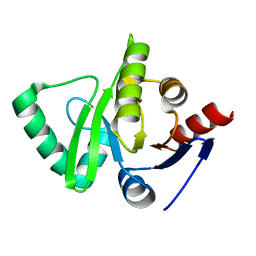 | | Crystal structure of HCoV-229E X-domain | | Descriptor: | Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
7S6F
 
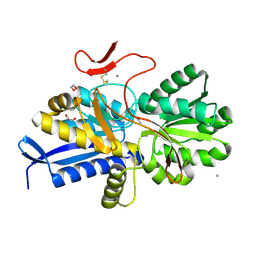 | | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative urea ABC transporter, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium
To Be Published
|
|
9GHZ
 
 | |
8IO3
 
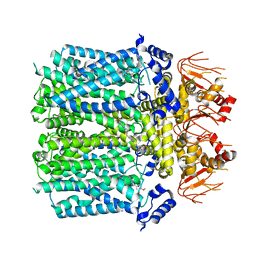 | | Cryo-EM structure of human HCN3 channel with cilobradine | | Descriptor: | 3-[[(3~{S})-1-[2-(3,4-dimethoxyphenyl)ethyl]piperidin-3-yl]methyl]-7,8-dimethoxy-2,5-dihydro-1~{H}-3-benzazepin-4-one, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure of human HCN3 channel with cilobradine
To Be Published
|
|
6DML
 
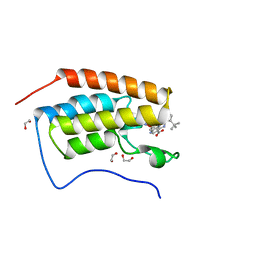 | | A multiconformer ligand model of 3,5 dimethylisoxaxole bound to the bromodomain of human BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(tert-butyl)phenyl)amino)-7-(3,5-dimethylisoxazol-4-yl)-6-methoxy-1,5-naphthyridine-3-carboxylic acid, Bromodomain-containing protein 4 | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
1M5I
 
 | |
3EKU
 
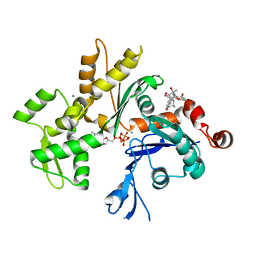 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
9G9U
 
 | | The structure of XynX, a NIF3 family protein from Geobacillus proteiniphilus T-6 | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase 1 type 2 homolog, ZINC ION | | Authors: | Hadad, N, Pomyalov, S, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2024-07-25 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of XynX, a NIF3 family protein from Geobacillus proteiniphilus T-6
To Be Published
|
|
3EMZ
 
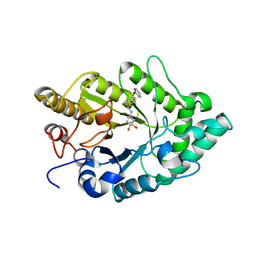 | | Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative | | Descriptor: | (1S,2S,3R,6R)-6-[(4-phenoxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
6SR3
 
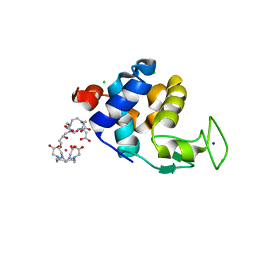 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 62 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6SR0
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: single colour reference data | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6D6L
 
 | |
2OX5
 
 | | The SoxYZ complex of Paracoccus pantotrophus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Bruno, S, Sauve, V, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2007-02-19 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The SoxYZ Complex Carries Sulfur Cycle Intermediates on a Peptide Swinging Arm.
J.Biol.Chem., 282, 2007
|
|
