7M4M
 
 | |
7MQY
 
 | | Zebrafish CNTN4 APLP2 complex | | Descriptor: | Fusion protein of CNTN4 and APLP2 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRO
 
 | | Zebrafish CNTN4 FN1-FN3 domains | | Descriptor: | Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRS
 
 | | Zebrafish CNTN4 APPb complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-09 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRM
 
 | | Chicken CNTN3 APP complex | | Descriptor: | Fusion protein of Chicken CNTN3 FN1-FN2 domains and Amyloid-beta A4 protein,Amyloid-beta A4 protein, SULFATE ION | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRK
 
 | | Chicken CNTN4 APP complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4, DI(HYDROXYETHYL)ETHER | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRN
 
 | | Mouse CNTN5 APP complex | | Descriptor: | Contactin-5, N-APP | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7XJC
 
 | | Crystal structure of bacteriorhodopsin in the ground and K states after green laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
5W1W
 
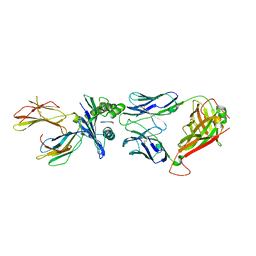 | | Structure of the HLA-E-VMAPRTLVL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-10-04 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
4ZOZ
 
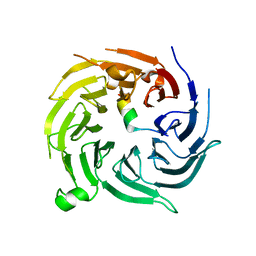 | |
5W69
 
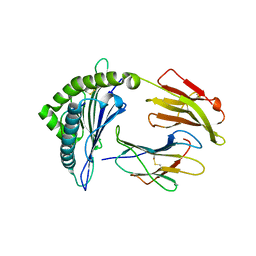 | | HLA-C*06:02 presenting ARFNDLRFV | | Descriptor: | ALA-ARG-PHE-ASN-ASP-LEU-ARG-PHE-VAL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
4ZK3
 
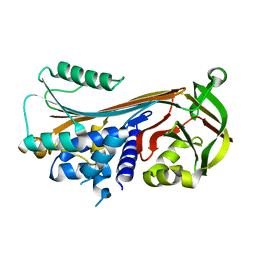 | |
5VL4
 
 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
5VD8
 
 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-099, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-chloro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
7NMH
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-070 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) methanesulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7N6W
 
 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
7N6U
 
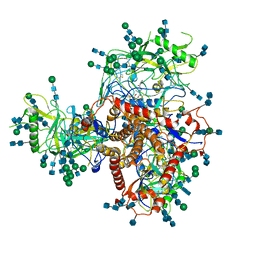 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
4ZEI
 
 | |
4ZOM
 
 | | RORgamma in complex with inverse agonist 4j. | | Descriptor: | N-{4-[3-(acetylamino)-1-(propan-2-yl)-1H-pyrazol-5-yl]-2-[(1R,5S)-3-azabicyclo[3.1.0]hex-3-yl]phenyl}-2-chloro-6-fluoro-N-methylbenzamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of novel pyrazole-containing benzamides as potent ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | Descriptor: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
5VQF
 
 | | Crystal Structure of pro-TGF-beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
4ZKJ
 
 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated protein Cas1, GLYCEROL | | Authors: | Ka, D, Bae, E. | | Deposit date: | 2015-04-30 | | Release date: | 2016-01-13 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Streptococcus pyogenes Cas1 and Its Interaction with Csn2 in the Type II CRISPR-Cas System
Structure, 24, 2016
|
|
4ZN0
 
 | |
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
4ZEC
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocin 84 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
