7VPR
 
 | |
5U6O
 
 | |
7V4Q
 
 | |
7K9H
 
 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7KC5
 
 | | X-ray structure of Lfa-1 I domain in complex with BMS-68852 collected at 273 K | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7K9J
 
 | |
7KC3
 
 | | X-ray structure of Lfa-1 I domain collected at 273 K | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC6
 
 | | X-ray structure of Lfa-1 I domain in complex with Lovastatin collected at 273 K | | Descriptor: | Integrin alpha-L, LOVASTATIN, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7VKA
 
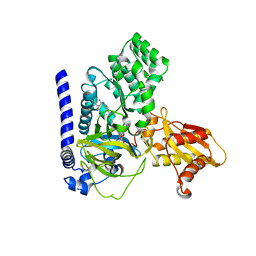 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5TSR
 
 | |
5TT2
 
 | |
5U1T
 
 | |
7VGN
 
 | | Crystal structure of CmnC | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CmnC, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7VKF
 
 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y, Nishiya, Y, Ito, K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VGL
 
 | | Crystal structure of CmnC | | Descriptor: | ACETATE ION, CmnC | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
5U9K
 
 | |
5TOA
 
 | |
5TPA
 
 | | Structure of the human GluN1/GluN2A LBD in complex with compound 9 (GNE3500) | | Descriptor: | (1R,2R)-2-(2-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-7-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-6-yl)cyclopropane-1-carbonitrile, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7VKD
 
 | |
7L32
 
 | |
5U1S
 
 | |
7WBK
 
 | |
7WBM
 
 | | Crystal structure of Legionella pneumophila effector protein Lpg0081 | | Descriptor: | Lpg0081, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Lee, J, Kim, H, Oh, B.H. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible modification of mitochondrial ADP/ATP translocases by paired Legionella effector proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W79
 
 | | Heme exporter HrtBA in complex with Mn-AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, Putative ABC transport system integral membrane protein, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
