7Q7L
 
 | |
7Q7I
 
 | |
7Q7W
 
 | |
6Q5S
 
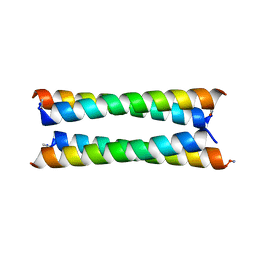 | | Crystal structure of a de novo designed antiparallel four-helix coiled coil apCC-Tet | | Descriptor: | apCC-Tet | | Authors: | Beesley, J.L, Guto, G.R, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
4WVL
 
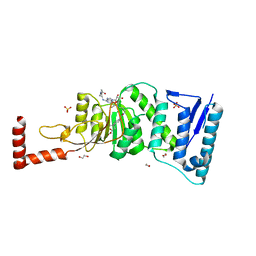 | | Structure-Guided DOT1L Probe Optimization by Label-Free Ligand Displacement | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Xu, X, Dhe-Paganon, S, Blacklow, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Guided DOT1L Probe Optimization by Label-Free Ligand Displacement.
Acs Chem.Biol., 10, 2015
|
|
4WKS
 
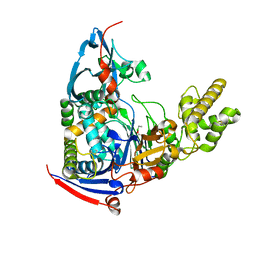 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, ethylboronic acid | | Authors: | Wu, R, Clevenger, D.K, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
8R3F
 
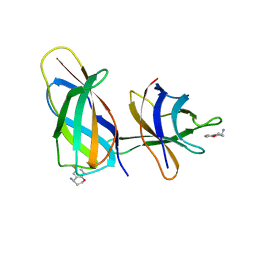 | | C-terminal Rel-homology Domain of NFAT1 | | Descriptor: | (4~{S})-6-fluoranyl-3,4-dihydro-2~{H}-chromen-4-amine, Nuclear factor of activated T-cells, cytoplasmic 2 | | Authors: | Zak, K.M, Boettcher, J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ligandability assessment of the C-terminal Rel-homology domain of NFAT1.
Arch Pharm, 357, 2024
|
|
4WK0
 
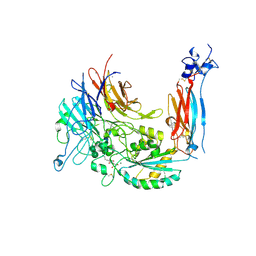 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARG-GLY-ASP, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WJK
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WKV
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, trihydroxy(octyl)borate(1-) | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1434 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
7CFO
 
 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
4WK2
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WKU
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, hexyl(trihydroxy)borate(1-) | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
5WQX
 
 | | Covalent bond formation of synthetic ligand with hPPARg-LBD | | Descriptor: | 2-[E-(E-2-oxidanylidenehexadec-5-enylidene)amino]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
4WK4
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-CYS-ARG-GLY-ASP-GLY-TRP-CYS, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WKT
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | 1-BUTANE BORONIC ACID, Acyl-homoserine lactone acylase PvdQ, GLYCEROL | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
8K1L
 
 | | Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in cation-free E2P form | | Descriptor: | Na+,K+-ATPase alpha2KK, Na+,K+-ATPase beta2, TETRAFLUOROALUMINATE ION | | Authors: | Abe, K, Artigas, P. | | Deposit date: | 2023-07-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A Na pump with reduced stoichiometry is up-regulated by brine shrimp in extreme salinities.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5X21
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | Descriptor: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zhang, Y, Ebright, R. | | Deposit date: | 2017-01-29 | | Release date: | 2017-07-05 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
2XK3
 
 | | Structure of Nek2 bound to Aminopyrazine compound 35 | | Descriptor: | 4-[3-AMINO-6-(3-ETHYLTHIOPHEN-2-YL)PYRAZIN-2-YL]CYCLOHEXANE-1-CARBOXYLIC ACID, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
5X22
 
 | |
4N7F
 
 | |
2FAR
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with dATP and Manganese | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6Q5I
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24E | | Descriptor: | AMMONIUM ION, CC-Hex*-L24E, SULFATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5L
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24H | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24H | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5P
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-II | | Descriptor: | CC-Hex*-II, GLYCEROL, PHOSPHATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
