5RUG
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000038389 | | Descriptor: | NAPHTHYLOXYACETIC ACID, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUT
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000161958 | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4FYJ
 
 | |
5RV8
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000039575 | | Descriptor: | 6-methylpyridine-3-carboxamide, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVP
 
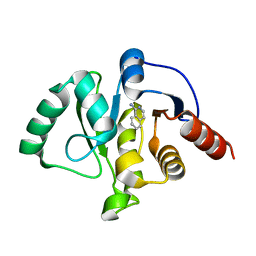 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000154817 | | Descriptor: | ISOQUINOLIN-1-AMINE, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4GKS
 
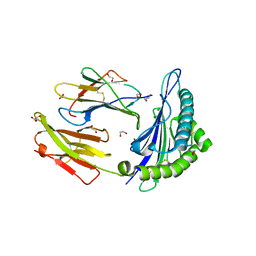 | | A2-MHC Complex carrying FLTGIGIITV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, FLT Cognate peptide, ... | | Authors: | Sewell, A.K, Rizkallah, P.J, Cole, D.K, Wooldridge, L, Price, D.A. | | Deposit date: | 2012-08-13 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | T-cell Receptor-optimized Peptide Skewing of the T-cell Repertoire Can Enhance Antigen Targeting.
J.Biol.Chem., 287, 2012
|
|
2R84
 
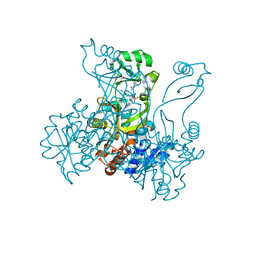 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP and AICAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
4FSL
 
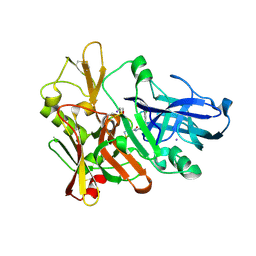 | |
3EMS
 
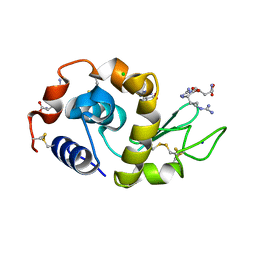 | | Effect of Ariginine on lysozyme | | Descriptor: | ARGININE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Matsuura, T, Yamaguhi, H. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Effect of Ariginine on lysozyme
to be published
|
|
3EN1
 
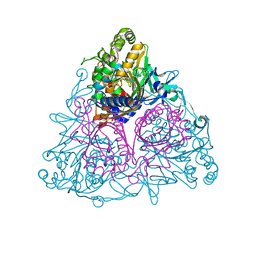 | | Crystal structure of Toluene 2,3-Dioxygenase | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Benzene 1,2-dioxygenase subunit alpha, Benzene 1,2-dioxygenase subunit beta, ... | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-09-25 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4FUK
 
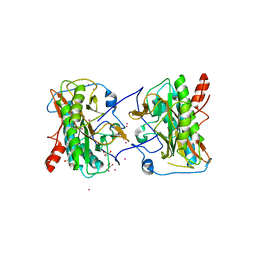 | | Aminopeptidase from Trypanosoma brucei | | Descriptor: | GLYCEROL, Methionine aminopeptidase, UNKNOWN ATOM OR ION, ... | | Authors: | El Bakkouri, M, Tempel, W, Osman, K.T, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminpeptidase from Trypanosoma brucei (CASP Target)
To be Published
|
|
2R8O
 
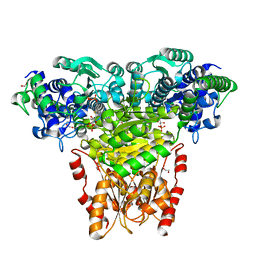 | | Transketolase from E. coli in complex with substrate D-xylulose-5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thia zol-3-ium-2-yl}-5-O-phosphono-D-xylitol, CALCIUM ION, ... | | Authors: | Wille, G, Asztalos, P, Weiss, M.S, Tittmann, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
2RBF
 
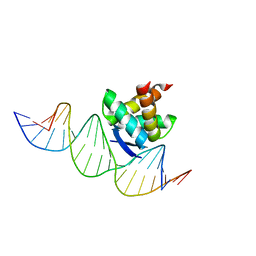 | | Structure of the ribbon-helix-helix domain of Escherichia coli PutA (PutA52) complexed with operator DNA (O2) | | Descriptor: | Bifunctional protein putA, DNA (5'-D(*DTP*DT*DTP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DCP*DAP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DGP*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DAP*DAP*DA)-3') | | Authors: | Tanner, J.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA.
J.Mol.Biol., 381, 2008
|
|
4FWP
 
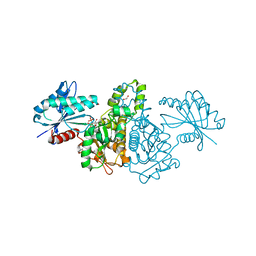 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
2RDR
 
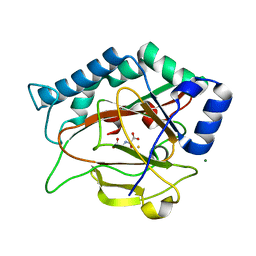 | | Crystal Structure of PtlH with Fe/oxalylglycine bound | | Descriptor: | 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, FE (III) ION, MAGNESIUM ION, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
2RCZ
 
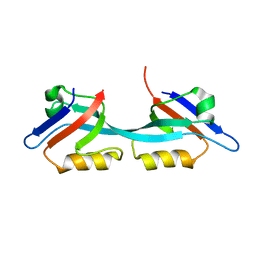 | | Structure of the second PDZ domain of ZO-1 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Lavie, A, Lye, M.F. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Domain swapping within PDZ2 is responsible for dimerization of ZO proteins.
J.Biol.Chem., 282, 2007
|
|
2RFE
 
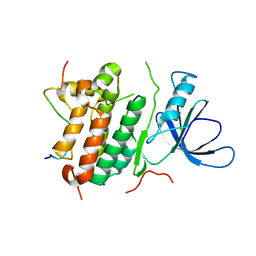 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
3E9R
 
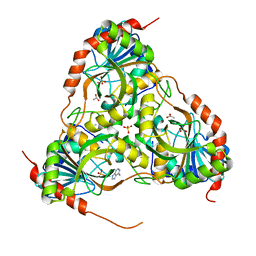 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with adenine | | Descriptor: | ACETATE ION, ADENINE, DIMETHYL SULFOXIDE, ... | | Authors: | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2RHO
 
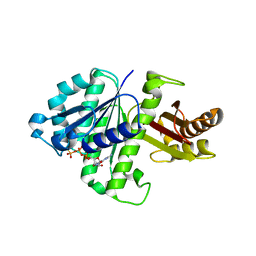 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ NCS Dimer with Bound GDP and GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell Division Protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RIX
 
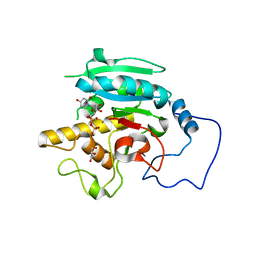 | | B-specific-1,3-galactosyltransferase)(GTB) + UDP | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ9
 
 | |
4FUR
 
 | |
2RID
 
 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with Allyl 7-O-carbamoyl-L-glycero-D-manno-heptopyranoside | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, prop-2-en-1-yl 7-O-carbamoyl-L-glycero-alpha-D-manno-heptopyranoside | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
5RS8
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000001601 | | Descriptor: | N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
