8PDO
 
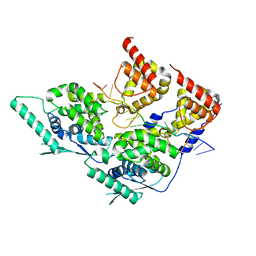 | | Local refinement of dimeric human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
9G02
 
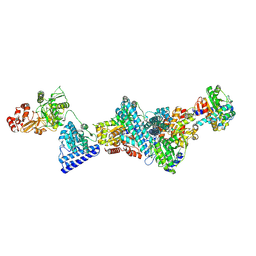 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (composite structure, semi-extended state) | | Descriptor: | CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase/acetyl-CoA synthase beta subunit, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
8PDL
 
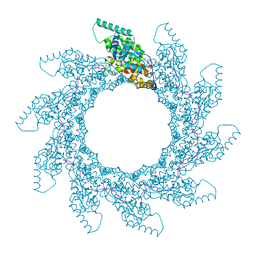 | | 10-mer ring of human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDS
 
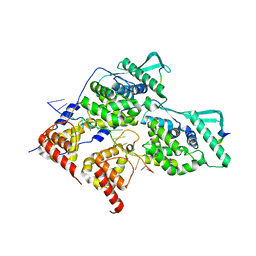 | | Local refinement of dimeric HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
9G01
 
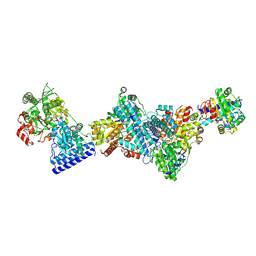 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (composite structure, closed and CO-bound state) | | Descriptor: | CARBON MONOXIDE, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase/acetyl-CoA synthase beta subunit, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9G00
 
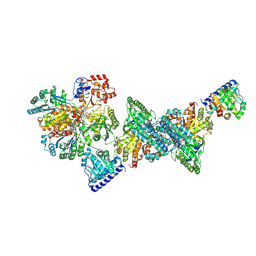 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (composite structure, class 3Cb) | | Descriptor: | Acetyl-CoA decarbonylase/synthase complex subunit delta, CO-methylating acetyl-CoA synthase, COBALAMIN, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9FZZ
 
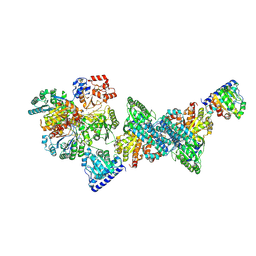 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (composite structure, class 3B) | | Descriptor: | Acetyl-CoA decarbonylase/synthase complex subunit delta, CO-methylating acetyl-CoA synthase, COBALAMIN, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9CRE
 
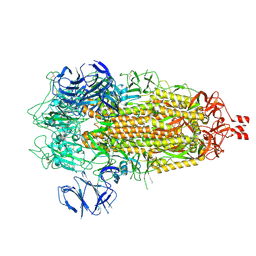 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9G03
 
 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with ferredoxin (Clostridium autoethanogenum) | | Descriptor: | CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase/acetyl-CoA synthase beta subunit, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9CRI
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Mu (B.1.621) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
2YDC
 
 | |
9FZY
 
 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (composite structure, class 3A) | | Descriptor: | Acetyl-CoA decarbonylase/synthase complex subunit delta, CO-methylating acetyl-CoA synthase, COBALAMIN, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
2Y6H
 
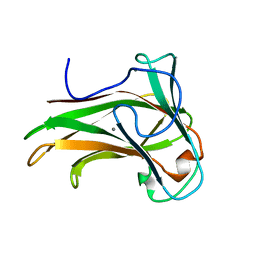 | | X-2 L110F CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
5BWY
 
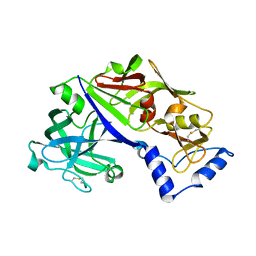 | | Structure of proplasmepsin II from Plasmodium falciparum, Space Group P43212 | | Descriptor: | Plasmepsin-2 | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2015-06-08 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4ZXO
 
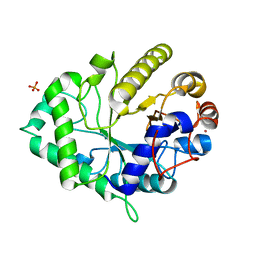 | | The structure of a GH26 beta-mannanase from Bacteroides ovatus, BoMan26A. | | Descriptor: | Glycosyl hydrolase family 26, PHOSPHATE ION, POTASSIUM ION | | Authors: | Bagenholm, V, Aurelius, O, Logan, D.T, Bouraoui, H, Stalbrand, H. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galactomannan Catabolism Conferred by a Polysaccharide Utilization Locus of Bacteroides ovatus: ENZYME SYNERGY AND CRYSTAL STRUCTURE OF A beta-MANNANASE.
J. Biol. Chem., 292, 2017
|
|
5CC2
 
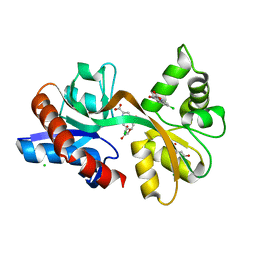 | | STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE IONOTROPIC GLUTAMATE RECEPTOR-LIKE GLUD2 IN COMPLEX WITH 7-CKA | | Descriptor: | 7-Chlorokynurenic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors.
Mol.Pharmacol., 89, 2016
|
|
2ZB0
 
 | | Crystal structure of P38 in complex with biphenyl amide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(3-cyanophenyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)-4-biphenylcarboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
9DTR
 
 | |
2FVV
 
 | | Human Diphosphoinositol polyphosphate phosphohydrolase 1 | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hallberg, B.M, Kursula, P, Ogg, D, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of human diphosphoinositol phosphatase 1
Proteins, 77, 2009
|
|
5CBS
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid at 1.8A resolution | | Descriptor: | (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
3B7T
 
 | | [E296Q]LTA4H in complex with Arg-Ala-Arg substrate | | Descriptor: | IMIDAZOLE, Leukotriene A-4 hydrolase, RAR peptide, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
9EVD
 
 | | In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Dietrich, L, Agip, A.N.A, Kuehlbrandt, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | In situ structure and rotary states of mitochondrial ATP synthase in whole Polytomella cells.
Science, 385, 2024
|
|
5NNS
 
 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
2H1W
 
 | | Crystal structure of the His183Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | FE (II) ION, Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
3B7S
 
 | | [E296Q]LTA4H in complex with RSR substrate | | Descriptor: | ACETIC ACID, GLYCEROL, Leukotriene A-4 hydrolase, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
