4UVY
 
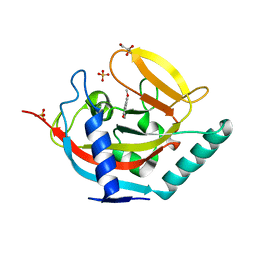 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methoxy-1,2- dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4V02
 
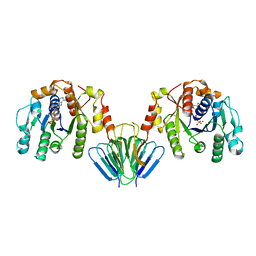 | | MinC:MinD cell division protein complex, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROBABLE SEPTUM SITE-DETERMINING PROTEIN MINC, ... | | Authors: | Ghosal, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
5LYB
 
 | |
6NDZ
 
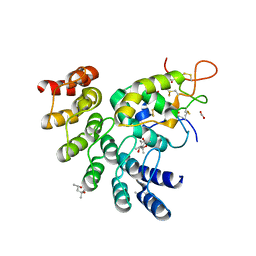 | | Designed repeat protein in complex with Fz8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ACETIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4V1P
 
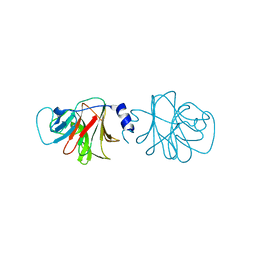 | | BTN3 Structure | | Descriptor: | BUTYROPHILIN SUBFAMILY 3 MEMBER A1 | | Authors: | James, L.C. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Activation of Human Gammadelta T Cells by Cytosolic Interactions of Btn3A1 with Soluble Phosphoantigens and the Cytoskeletal Adaptor Periplakin.
J.Immunol., 194, 2015
|
|
6NEJ
 
 | | scoulerine 9-O-methyltransferase from Thalictrum flavum complexed (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol and with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, (S)-scoulerine 9-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Valentic, T.R, Smolke, C.D, Payne, J.T. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of the scoulerine 9-O methyltransferase from Thalictrum flavum.
To Be Published
|
|
4US1
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | (3S)-3-[3-(aminomethyl)phenyl]-1-ethylpyrrolidine-2,5-dione, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
6NEX
 
 | | Fab fragment of anti-cocaine antibody h2E2 | | Descriptor: | ACETATE ION, Anitgen binding fragment light chain, Antigen binding fragment heavy chain, ... | | Authors: | Pokkuluri, P.R, Tan, K. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4UWU
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 7) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
4V0H
 
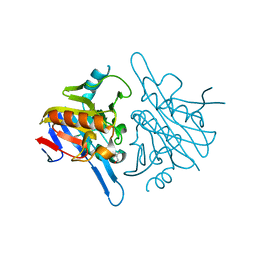 | | Human metallo beta lactamase domain containing protein 1 (hMBLAC1) | | Descriptor: | FE (III) ION, GLYCEROL, METALLO-BETA-LACTAMASE DOMAIN-CONTAINING PROTEIN 1 1 | | Authors: | Pettinati, I, McDonough, M.A, Brem, J, Schofield, C.J. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Biosynthesis of histone messenger RNA employs a specific 3' end endonuclease.
Elife, 7, 2018
|
|
6NF6
 
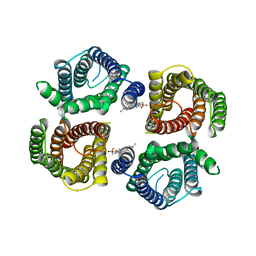 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NG4
 
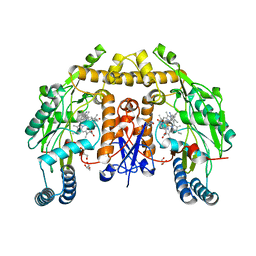 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with (R)-6-(3-fluoro-5-(2-(pyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R)-pyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
6NFW
 
 | |
4USZ
 
 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
6NBP
 
 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
6NBU
 
 | |
4V1W
 
 | |
6NKJ
 
 | | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid.
To Be Published
|
|
4V16
 
 | | KlHsv2 with loop 6CD replaced by a Gly-Ser linker | | Descriptor: | SVP1-LIKE PROTEIN 2 | | Authors: | Busse, R.A, Scacioc, A, Krick, R, Perez-Lara, A, Thumm, M, Kuhnel, K. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Proppin-Phosphoinositide Binding and Role of Loop 6Cd in Proppin-Membrane Binding.
Biophys.J., 108, 2015
|
|
4UQS
 
 | |
6NMC
 
 | | CryoEM structure of the LbCas12a-crRNA-2xAcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4UV0
 
 | | Structure of a semisynthetic phosphorylated DAPK | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, TRIETHYLENE GLYCOL | | Authors: | de Diego, I, Rios, P, Meyer, C, Koehn, M, Wilmanns, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular Mechanisms Behind Dapk Regulation: How the Phosphorylation Activity Switch Works
To be Published
|
|
4UVR
 
 | | Binding mode, selectivity and potency of N-indolyl-oxopyridinyl-4- amino-propanyl-based inhibitors targeting Trypanosoma cruzi CYP51 | | Descriptor: | Nalpha-{4-[4-(5-chloro-2-methylphenyl)piperazin-1-yl]-2-fluorobenzoyl}-N-pyridin-4-yl-D-tryptophanamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-DEMETHYLASE, ... | | Authors: | Vieira, D.F, Choi, J.Y, Calvet, C.M, Gut, J, Kellar, D, Siqueira-Neto, J.L, Johnston, J.B, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
6NN7
 
 | | The structure of human liver pyruvate kinase, hLPYK-GGG | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
