5FUH
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
6N0R
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 | | Descriptor: | 1,2-ETHANEDIOL, 4-(methylamino)benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
4RZ3
 
 | | Crystal structure of the MinD-like ATPase FlhG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Site-determining protein | | Authors: | Schuhmacher, J.S, Bange, G. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6Q4A
 
 | | CDK2 in complex with FragLite14 | | Descriptor: | 5-iodanylpyrimidine, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7ZRQ
 
 | | 1.68 Angstrom crystal structure of Ca/CaM-E140G:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
6Q4I
 
 | | CDK2 in complex with FragLite35 | | Descriptor: | 2-[4-[(2-oxidanylidene-3~{H}-pyridin-4-yl)oxy]phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4C
 
 | | CDK2 in complex with FragLite16 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4F
 
 | | CDK2 in complex with FragLite32 | | Descriptor: | Cyclin-dependent kinase 2, PYRIDINE-2,6-DIAMINE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6FX3
 
 | | crystal structure of Pholiota squarrosa lectin in complex with a dodecasaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, lectin | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6Q57
 
 | |
4RKH
 
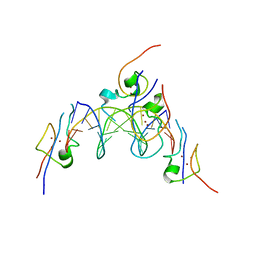 | | Structure of the MSL2 CXC domain bound with a specific MRE sequence | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*CP*TP*CP*GP*CP*TP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*AP*GP*CP*GP*AP*GP*AP*TP*GP*GP*AP*T)-3'), E3 ubiquitin-protein ligase msl-2, ... | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
6N4Q
 
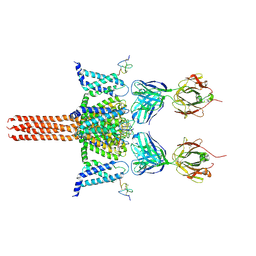 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
5FRQ
 
 | |
6G6K
 
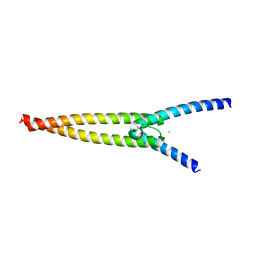 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | CHLORIDE ION, Myc proto-oncogene protein, Protein max | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
6N0B
 
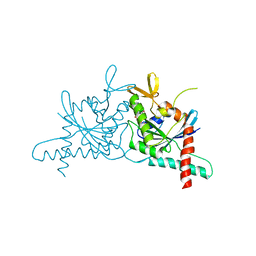 | | Structure of GTPase Domain of Human Septin 7 at High Resolution | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin-7 | | Authors: | Brognara, G, Pereira, H.M, Brandao-Neto, J, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2018-11-07 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Revisiting SEPT7 and the slippage of beta-strands in the septin family.
J.Struct.Biol., 207, 2019
|
|
6G8W
 
 | |
6QDD
 
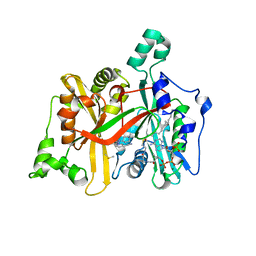 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000105 | | Descriptor: | 2-[(2~{S})-1-[2-[methyl-(1-methylpiperidin-4-yl)amino]thieno[3,2-d]pyrimidin-4-yl]-2,3-dihydropyrrol-2-yl]ethanenitrile, DIMETHYL SULFOXIDE, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDC
 
 | |
6N8I
 
 | |
6N2K
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[2-{[(2R)-1-(dimethylamino)propan-2-yl]oxy}-7-(3-hydroxynaphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
7ZPC
 
 | | CDK2 in complex 9K-DOS | | Descriptor: | Cyclin-dependent kinase 2, ~{N}-(1-methylpyrazol-4-yl)-5-phenyl-pyrazolo[1,5-a]pyrimidine-7-carboxamide | | Authors: | Watt, J.E, Martin, M.P, Noble, M.E.N. | | Deposit date: | 2022-04-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Build-Couple-Transform: A Paradigm for Lead-like Library Synthesis with Scaffold Diversity.
J.Med.Chem., 65, 2022
|
|
6QGG
 
 | | Structure of human Bcl-2 in complex with analogue of ABT-737 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, [(3~{R})-3-[[4-[[4-[4-[[2-(4-chlorophenyl)phenyl]methyl]piperazin-1-yl]phenyl]carbonylsulfamoyl]-2-nitro-phenyl]amino]-4-phenylsulfanyl-butyl]-(2-hydroxy-2-oxoethyl)-dimethyl-azanium | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6MVK
 
 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{(4S)-3-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-2-oxo-1,3-oxazolidin-4-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
8WZU
 
 | |
4S28
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound aminoimidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Fe | | Descriptor: | 1,4-BUTANEDIOL, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
