1M85
 
 | | Structure of Proteus mirabilis catalase for the native form | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Gouet, P, Jouve, H.-M, Dideberg, O. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Proteus mirabilis PR catalase with and without bound NADPH.
J.Mol.Biol., 249, 1995
|
|
4R68
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 31 | | Descriptor: | (1S)-1-phenylethyl (4-chloro-3-{[(4S)-4-(2,6-dichlorophenyl)-2-hydroxy-6-oxocyclohex-1-en-1-yl]sulfanyl}phenyl)acetate, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
6AH5
 
 | | Structure of human P2X3 receptor in complex with ATP and Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.819 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4JDB
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-005 | | Descriptor: | 5-[2-(7-methoxynaphthalen-2-yl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-005
To be Published
|
|
6U4J
 
 | | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | | Descriptor: | 5-{[(1S)-1-(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]amino}-1-methyl-6-oxo-1,6-dihydropyridine-2-carbonitrile, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-08-25 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6EKQ
 
 | | Crystal structure of the human BRPF1 bromodomain complexed with BZ054 in space group C2 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[4-[[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]methyl]-1,3-thiazol-2-yl]-2,4-dimethyl-1,3-oxazole-5-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5ZWJ
 
 | | Crystal structure of EGFR 675-1022 T790M/C797S/V948R in complex with EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor | | Authors: | Zhao, P, Yun, C.H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of EGFR T790M/C797S/V948R in complex with EAI045.
Biochem. Biophys. Res. Commun., 502, 2018
|
|
6A5E
 
 | | Crystal structure of plant peptide RALF23 in complex with FER and LLG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GPI-anchored protein LLG2, ... | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2018-06-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.766 Å) | | Cite: | Mechanisms of RALF peptide perception by a heterotypic receptor complex.
Nature, 572, 2019
|
|
5OWN
 
 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
3VDR
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase, prepared in the presence of the substrate D-3-hydroxybutyrate and NAD(+) | | Descriptor: | (3R)-3-hydroxybutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETIC ACID, ... | | Authors: | Hoque, M.M, Shimizu, S, Juan, E.C.M, Sato, Y, Hossain, M.T, Yamamoto, T, Imamura, S, Amano, H, Suzuki, K, Sekiguchi, T, Tsunoda, M, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7BI9
 
 | | PI3KC2a core in complex with PIK90 | | Descriptor: | 1,2-ETHANEDIOL, N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha,Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1LY3
 
 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5X24
 
 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5DTV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dimethyl-substituted, 3,4-diarylthiophene dioxide core ligand | | Descriptor: | 3,4-bis(4-hydroxy-2-methylphenyl)-1H-1lambda~6~-thiophene-1,1-dione, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
9IHT
 
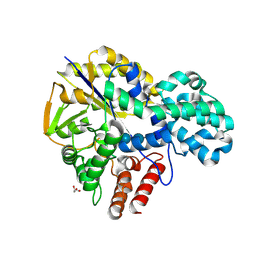 | | Crystal structure of GH57 family amylopullulanase from Aquifex aeolicus in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 57 N-terminal domain-containing protein | | Authors: | Zhu, Z.M, Wang, W.W, Yu, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | The crystal structure of GH57 family amylopullulanase reveals its dual binding pockets sharing the same catalytic dyad.
Commun Biol, 8, 2025
|
|
9HH4
 
 | | Crystal structure of the family S1_19 carrageenan sulfatase ZgCgsA from Zobellia galactanivorans in complex with hybrid b-k-neocarratetraose | | Descriptor: | 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Chevenier, A, Czjzek, M, Michel, G, Ficko-Blean, E. | | Deposit date: | 2024-11-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, function and catalytic mechanism of the carrageenan-sulfatases from the marine bacterium Zobellia galactanivorans Dsij T.
Carbohydr Polym, 358, 2025
|
|
6AN1
 
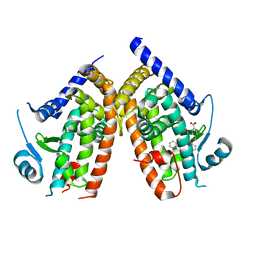 | | Crystal structure of the complex between PPARgamma LBD and the ligand AM-879 | | Descriptor: | 4-({2-[(1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)methyl]phenoxy}methyl)benzoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Veras, H, Figueira, A.C, le Maire, A. | | Deposit date: | 2017-08-11 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Screening for PPAR Non-Agonist Ligands Followed by Characterization of a Hit, AM-879, with Additional No-Adipogenic and cdk5-Mediated Phosphorylation Inhibition Properties.
Front Endocrinol (Lausanne), 9, 2018
|
|
6AH4
 
 | | Structure of human P2X3 receptor in complex with ATP and Ca2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
1AHZ
 
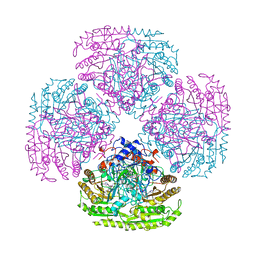 | |
6RQF
 
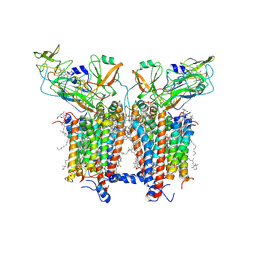 | | 3.6 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Spinacia oleracea with natively bound thylakoid lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Malone, L.A, Qian, P, Mayneord, G.E, Hitchcock, A, Farmer, D, Thompson, R, Swainsbury, D.J.K, Ranson, N, Hunter, C.N, Johnson, M.P. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-13 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structure of the spinach cytochrome b6f complex at 3.6 angstrom resolution.
Nature, 575, 2019
|
|
1MNP
 
 | | MANGANESE PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Sundaramoorthy, M, Poulos, T.L. | | Deposit date: | 1995-01-27 | | Release date: | 1995-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preliminary crystallographic analysis of manganese peroxidase from Phanerochaete chrysosporium.
J.Mol.Biol., 238, 1994
|
|
1AA6
 
 | | REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
8XKL
 
 | | Structure of ACPII-CCPII from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
6AT2
 
 | | E. coli phosphoenolpyruvate carboxykinase G209N mutant bound to thiosulfate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-27 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
