6TSI
 
 | |
5ZTO
 
 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with D3003 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-{trans-4-[3-(2-chlorophenyl)-7-{[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]cyclohexyl}propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2018-05-04 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystal structure of EGFR 696-1022 T790M/C797S in complex with D3003
To Be Published
|
|
6EER
 
 | | Structure of glycine-bound GoxA from Pseudoalteromonas luteoviolacea | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GoxA, ... | | Authors: | Yukl, E.T, Avalos, D. | | Deposit date: | 2018-08-15 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Spectroscopic Characterization of a Product Schiff Base Intermediate in the Reaction of the Quinoprotein Glycine Oxidase, GoxA.
Biochemistry, 58, 2019
|
|
5ZRF
 
 | | Crystal structure of human topoisomerase II beta in complex with 5-iodouridine-containing-DNA and etoposide in space group p21 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Chen, S.F, Wang, Y.R, Wu, C.C, Chan, N.L. | | Deposit date: | 2018-04-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the gating of DNA passage by the topoisomerase II DNA-gate.
Nat Commun, 9, 2018
|
|
5CI1
 
 | | Ribonucleotide reductase Y122 2,3-F2Y variant | | Descriptor: | CHLORIDE ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
4F4M
 
 | |
5H4I
 
 | |
9GBI
 
 | | Cryo-EM structure of Arabidopsis thaliana PSI-LHCI wild-type | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Capaldi, S, Chaves-Sanjuan, A, Bonnet, D.M.V, Bassi, R. | | Deposit date: | 2024-07-31 | | Release date: | 2025-08-13 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural determinants for red-shifted absorption in higher-plants Photosystem I.
New Phytol., 2025
|
|
9GC2
 
 | | Cryo-EM structure of Arabidopsis thaliana PSI-LHCI- a603-NH mutant | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Capaldi, S, Chaves-Sanjuan, A, Bonnet, D.M.V, Bassi, R. | | Deposit date: | 2024-08-01 | | Release date: | 2025-08-13 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural determinants for red-shifted absorption in higher-plants Photosystem I.
New Phytol., 2025
|
|
9M4F
 
 | | Photosystem I from the eukaryotic filamentous algae | | Descriptor: | (1~{R},3~{S})-6-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{Z},17~{E})-16-(hydroxymethyl)-3,7,12-trimethyl-18-[(1~{S},4~{S},6~{R})-2,2,6-trimethyl-4-oxidanyl-7-oxabicyclo[4.1.0]heptan-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenylidene]-1,5,5-trimethyl-cyclohexane-1,3-diol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Shao, R.Q, Pan, X.W. | | Deposit date: | 2025-03-04 | | Release date: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Architecture of photosystem I-light-harvesting complex from the eukaryotic filamentous yellow-green alga Tribonema minus.
J Integr Plant Biol, 2025
|
|
6AHE
 
 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ternary complex formation of AFN-1252 with Acinetobacter baumannii FabI and NADH: Crystallographic and biochemical studies.
Chem.Biol.Drug Des., 96, 2020
|
|
4O8U
 
 | | Structure of PF2046 | | Descriptor: | Uncharacterized protein PF2046 | | Authors: | Su, J, Liu, Z.-J. | | Deposit date: | 2013-12-30 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Crystal structure of a novel non-Pfam protein PF2046 solved using low resolution B-factor sharpening and multi-crystal averaging methods
Protein Cell, 1, 2010
|
|
8AX5
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029881 | | Descriptor: | (1~{R},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3,5,7(30),20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
8ZTU
 
 | | 70S ribosome arrested by PepNL | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Ando, Y, Kobo, A, Nureki, O, Taguchi, H, Itoh, Y, Chadani, Y. | | Deposit date: | 2024-06-07 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A mini-hairpin shaped nascent peptide blocks translation termination by a distinct mechanism.
Nat Commun, 16, 2025
|
|
8ZTV
 
 | | 70S ribosome arrested by PepNL with RF2 | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Ando, Y, Kobo, A, Nureki, O, Taguchi, H, Itoh, Y, Chadani, Y. | | Deposit date: | 2024-06-07 | | Release date: | 2025-03-19 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A mini-hairpin shaped nascent peptide blocks translation termination by a distinct mechanism.
Nat Commun, 16, 2025
|
|
5GLX
 
 | | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
6HOL
 
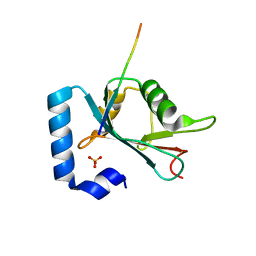 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6WYF
 
 | |
8AX6
 
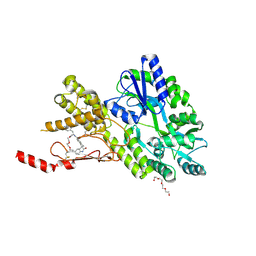 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029882 | | Descriptor: | (1~{S},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
8AX7
 
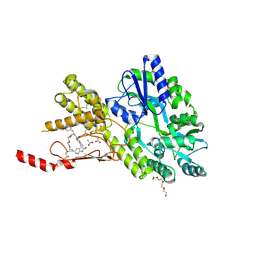 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0031448 | | Descriptor: | (1~{S},10~{R},20~{E})-10-[(1,7-dimethylindazol-5-yl)methyl]-12-methyl-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, ACETATE ION, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
8IQT
 
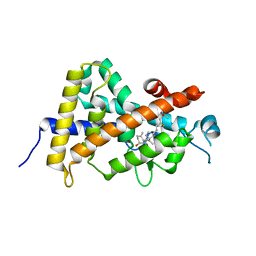 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (23R)-F-25(OH)D3 | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},4~{R})-4-fluoranyl-6-methyl-6-oxidanyl-heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Vitamin D3 receptor | | Authors: | Kakuda, S. | | Deposit date: | 2023-03-17 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (23R)-F-25(OH)D3
To Be Published
|
|
5K6U
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, IODIDE ION, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
6QNB
 
 | |
6EAV
 
 | |
6E64
 
 | |
