2G9F
 
 | | Crystal structure of intein-tagged mouse PNGase C-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | Descriptor: | GLYCEROL, SULFATE ION, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G9H
 
 | | Crystal Structure of Staphylococcal Enterotoxin I (SEI) in Complex with a Human MHC class II Molecule | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HLA class II histocompatibility antigen, ... | | Authors: | Fernandez, M.M, Guan, R, Malchiodi, E.L, Mariuzza, R.A. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule.
J.Biol.Chem., 281, 2006
|
|
2G9I
 
 | | Crystal structure of homolog of F420-0:gamma-Glutamyl Ligase from Archaeoglobus fulgidus Reveals a Novel Fold. | | Descriptor: | F420-0:gamma-glutamyl ligase | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-04-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an amide bond forming F(420):gamma-glutamyl ligase from Archaeoglobus fulgidus -- a member of a new family of non-ribosomal peptide synthases.
J.Mol.Biol., 372, 2007
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
2G9K
 
 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4-hydroxy-2',3,3',4',5-Pentachlorobiphenyl | | Descriptor: | 2',3,3',4',5-PENTACHLOROBIPHENYL-4-OL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-03-06 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
2G9L
 
 | |
2G9N
 
 | | Structure of the DEAD domain of Human eukaryotic initiation factor 4A, eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I | | Authors: | Hogbom, M, Ogg, D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-07 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | Descriptor: | antimicrobial peptide Latarcin 2a | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2006-03-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
2G9Q
 
 | | The crystal structure of the glycogen phosphorylase b- 1AB complex | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-D-ARABINITOL, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E, Kristiansen, M, Agius, L. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
2G9R
 
 | | The crystal structure of glycogen phosphorylase b in complex with (3R,4R,5R)-5-hydroxymethyl-1-(3-phenylpropyl)-piperidine-3,4-diol | | Descriptor: | (3R,4R,5R)-5-(HYDROXYMETHYL)-1-(3-PHENYLPROPYL)PIPERIDINE-3,4-DIOL, Glycogen phosphorylase, muscle form | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
2G9T
 
 | | Crystal structure of the SARS coronavirus nsp10 at 2.1A | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Yang, H, Sun, F, Rao, Z. | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2G9U
 
 | | The crystal structure of glycogen phosphorylase in complex with (3R,4R,5R)-5-hydroxymethyl-1-(3-phenylpropyl)-piperidine-3,4-diol and phosphate | | Descriptor: | (3R,4R,5R)-5-(HYDROXYMETHYL)-1-(3-PHENYLPROPYL)PIPERIDINE-3,4-DIOL, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
2G9V
 
 | | The crystal structure of glycogen phosphorylase in complex with (3R,4R,5R)-5-hydroxymethylpiperidine-3,4-diol and phosphate | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
2G9W
 
 | | Crystal Structure of Rv1846c, a Putative Transcriptional Regulatory Protein of Mycobacterium Tuberculosis | | Descriptor: | CHLORIDE ION, conserved hypothetical protein | | Authors: | Saul, F.A, Haouz, A, Fiez-Vandal, C, Shepard, W, Alzari, P.M. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis
Mol.Microbiol., 71, 2009
|
|
2G9X
 
 | |
2G9Y
 
 | |
2G9Z
 
 | | Thiamin pyrophosphokinase from Candida albicans | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-(2-{[HYDROXY(PHOSPHONOAMINO)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-I UM, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Abergel, C, Santini, S, Monchois, V, Rousselle, T, Claverie, J.M, Bacterial targets at IGS-CNRS, France (BIGS) | | Deposit date: | 2006-03-07 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase.
Bmc Struct.Biol., 8, 2008
|
|
2GA0
 
 | | Variable Small Protein 1 of Borrelia turicatae (VspA or Vsp1) | | Descriptor: | NICKEL (II) ION, surface protein VspA | | Authors: | Lawson, C.L, Yung, B.H, Barbour, A.G, Zuckert, W.R. | | Deposit date: | 2006-03-07 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of neurotropism-associated variable surface protein 1 (Vsp1) of Borrelia turicatae.
J.Bacteriol., 188, 2006
|
|
2GA1
 
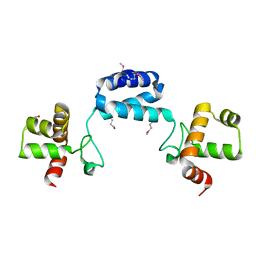 | |
2GA2
 
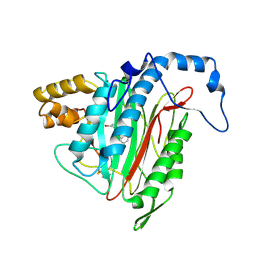 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2GA3
 
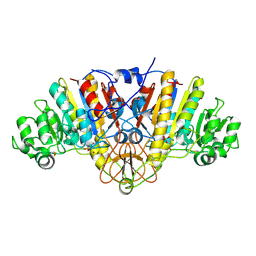 | |
2GA4
 
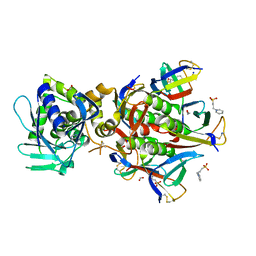 | | Stx2 with adenine | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ADENINE, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2006-03-07 | | Release date: | 2006-07-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of adenine to Stx2, the protein toxin from Escherichia coli O157:H7.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GA5
 
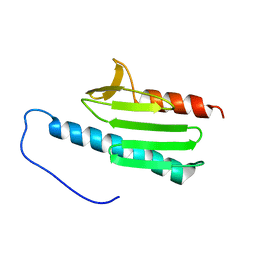 | | yeast frataxin | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | He, Y, Alam, S.L, Proteasa, S.V, Zhang, Y, Lesuisse, E, Dancis, A. | | Deposit date: | 2006-03-07 | | Release date: | 2006-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Yeast Frataxin Solution Structure, Iron Binding and Ferrochelatase Interaction
Biochemistry, 43, 2004
|
|
