1S14
 
 | | Crystal structure of Escherichia coli Topoisomerase IV ParE 24kDa subunit | | Descriptor: | NOVOBIOCIN, Topoisomerase IV subunit B | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
1N9G
 
 | | Mitochondrial 2-enoyl thioester reductase Etr1p/Etr2p heterodimer from Candida tropicalis | | Descriptor: | 2,4-dienoyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Torkko, J.M, Koivuranta, K.T, Kastaniotis, A.J, Airenne, T.T, Glumoff, T, Ilves, M, Hartig, A, Gurvitz, A, Hiltunen, J.K. | | Deposit date: | 2002-11-25 | | Release date: | 2003-12-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Candida tropicalis expresses two mitochondrial 2-enoyl thioester reductases that are able to form both homodimers and heterodimers.
J.Biol.Chem., 278, 2003
|
|
1S46
 
 | | Covalent intermediate of the E328Q amylosucrase mutant | | Descriptor: | amylosucrase, beta-D-glucopyranose | | Authors: | Jensen, M.H, Mirza, O, Albenne, C, Remaud-Simeon, M, Monsan, P, Gajhede, M, Skov, L.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea.
Biochemistry, 43, 2004
|
|
1GQ1
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
2JFQ
 
 | |
1H78
 
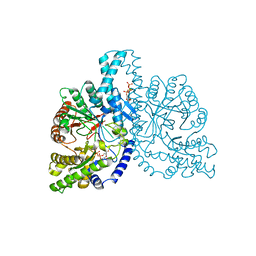 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DCTP. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, MAGNESIUM ION | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2001-07-04 | | Release date: | 2002-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
1H19
 
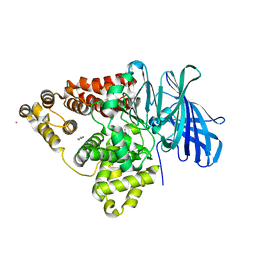 | | STRUCTURE OF [E271Q]LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2002-07-04 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Leukotriene A4 Hydrolase/Aminopeptidase, Glutamate 271 is a Catalyticresidue with Specific Roles in Two Distinct Enzyme Mechanisms
J.Biol.Chem., 277, 2002
|
|
1H6R
 
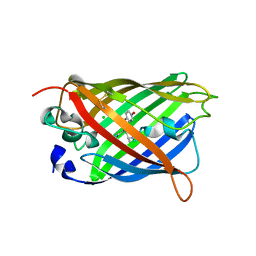 | | The oxidized state of a redox sensitive variant of green fluorescent protein | | Descriptor: | CHLORIDE ION, GREEN FLUORESCENT PROTEIN | | Authors: | Ostergaard, H, Henriksen, A, Hansen, F.G, Winther, J.R. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Shedding Light on Disulfide Bond Formation: Engineering a Redox Switch in Green Fluorescent Protein
Embo J., 20, 2001
|
|
1H7X
 
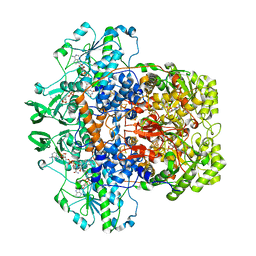 | | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Dihydropyrimidine Dehydrogenase, a Major Determinant of the Pharmacokinetics of the Anti-Cancer Drug 5-Fluorouracil
Embo J., 20, 2001
|
|
1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
1GZ9
 
 | | High-Resolution Crystal Structure of Erythrina cristagalli Lectin in Complex with 2'-alpha-L-Fucosyllactose | | Descriptor: | CALCIUM ION, ERYTHRINA CRISTA-GALLI LECTIN, MANGANESE (II) ION, ... | | Authors: | Svensson, C, Teneberg, S, Nilsson, C.L, Kjellberg, A, Schwarz, F.P, Sharon, N, Krengel, U. | | Deposit date: | 2002-05-17 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structures of Erythrina Cristagalli Lectin in Complex with Lactose and 2'-Alpha-L-Fucosyllactose and Correlation with Thermodynamic Binding Data
J.Mol.Biol., 321, 2002
|
|
1M1T
 
 | | Biosynthetic thiolase, Q64A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M4S
 
 | | Biosynthetic thiolase, Cys89 acetylated, unliganded form | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1LY8
 
 | | The crystal structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability and insight into modelling of protein structures | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Houborg, K, Harris, P, Poulsen, J.-C.N, Svendsen, A, Schneider, P, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N5A
 
 | | Crystal structure of the Murine class I Major Histocompatibility Complex of H-2DB, B2-Microglobulin, and A 9-Residue immunodominant peptide epitope gp33 derived from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Odeberg, J, Grufman, P, Sandberg, J.K, Levitsky, V, Karre, K, Sandalova, T, Schneider, G. | | Deposit date: | 2002-11-05 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of
gp33 Revealed by Comparative Crystal Structure Analyses.
Immunity, 17, 2002
|
|
1H75
 
 | | Structural basis for the thioredoxin-like activity profile of the glutaredoxin-like protein NrdH-redoxin from Escherichia coli. | | Descriptor: | GLUTAREDOXIN-LIKE PROTEIN NRDH | | Authors: | Stehr, M, Schneider, G, Aslund, F, Holmgren, A, Lindqvist, Y. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Thioredoxin-Like Activity Profile of the Glutaredoxin-Like Nrdh-Redoxin from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1H58
 
 | | STRUCTURE OF FERROUS HORSERADISH PEROXIDASE C1A | | Descriptor: | ACETATE ION, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-20 | | Release date: | 2002-06-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1HJS
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1LEL
 
 | | The avidin BCAP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, E-AMINO BIOTINYL CAPROIC ACID | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-10 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1SI8
 
 | | Crystal structure of E. faecalis catalase | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hakansson, K.O, Brugna, M, Tasse, L. | | Deposit date: | 2004-02-28 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of catalase from Enterococcus faecalis.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S7X
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
2JAC
 
 | |
1Q3I
 
 | | Crystal Structure of Na,K-ATPase N-domain | | Descriptor: | NICKEL (II) ION, Na,K-ATPase | | Authors: | Hakansson, K.O. | | Deposit date: | 2003-07-30 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystallographic Structure of Na,K-ATPase N-domain at 2.6 A Resolution
J.Mol.Biol., 332, 2003
|
|
3ICR
 
 | | Crystal structure of oxidized Bacillus anthracis CoADR-RHD | | Descriptor: | COENZYME A, Coenzyme A-Disulfide Reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
1GQ8
 
 | | Pectin methylesterase from Carrot | | Descriptor: | CACODYLATE ION, PECTINESTERASE | | Authors: | Johansson, K, El-Ahmad, M, Friemann, R, Jornvall, H, Markovic, O, Eklund, H. | | Deposit date: | 2001-11-20 | | Release date: | 2002-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Plant Pectin Methylesterase
FEBS Lett., 514, 2002
|
|
