1UF0
 
 | | Solution structure of the N-terminal DCX domain of human doublecortin-like kinase | | Descriptor: | Serine/threonine-protein kinase DCAMKL1 | | Authors: | Saito, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal DCX domain of human doublecortin-like kinase
To be Published
|
|
1UFZ
 
 | | Solution structure of HBS1-like domain in hypothetical protein BAB28515 | | Descriptor: | Hypothetical protein BAB28515 | | Authors: | He, F, Muto, Y, Hayami, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HBS1-like domain in hypothetical protein BAB28515
To be Published
|
|
3ARO
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure | | Descriptor: | Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARY
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
1Q6A
 
 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3ARX
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Propentofylline | | Descriptor: | 3-methyl-1-(5-oxohexyl)-7-propyl-3,7-dihydro-1H-purine-2,6-dione, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
1UEM
 
 | | Solution Structure of the First Fibronectin Type III domain of human KIAA1568 Protein | | Descriptor: | KIAA1568 Protein | | Authors: | Li, H, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the First Fibronectin Type III domain of human KIAA1568 Protein
To be Published
|
|
1UE9
 
 | | Solution structure of the fourth SH3 domain of human intersectin 2 (KIAA1256) | | Descriptor: | Intersectin 2 | | Authors: | Tochio, N, Kigawa, T, Koshiba, S, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-09 | | Release date: | 2003-11-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth SH3 domain of human intersectin 2 (KIAA1256)
To be Published
|
|
1UFL
 
 | | Crystal Structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Hori-Takemoto, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-31 | | Release date: | 2003-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
1UFF
 
 | | Solution structure of the first SH3 domain of human intersectin2 (KIAA1256) | | Descriptor: | Intersectin 2 | | Authors: | Kobayashi, N, Kigawa, T, Koshiba, S, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first SH3 domain of human intersectin2 (KIAA1256)
To be Published
|
|
1UG8
 
 | | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease | | Descriptor: | Poly(A)-specific Ribonuclease | | Authors: | Nagata, T, Muto, Y, Hayami, N, Uda, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease
To be Published
|
|
1Q8Z
 
 | | The apoenzyme structure of the yeast SR protein kinase, Sky1p | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, SR Protein Kinase, ... | | Authors: | Nolen, B, Ngo, J, Chakrabarti, S, Vu, D, Adams, J.A, Ghosh, G. | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Nucleotide-Induced Conformational Changes in the Saccharomyces cerevisiae SR Protein Kinase, Sky1p, Revealed by X-Ray Crystallography
Biochemistry, 42, 2003
|
|
1QWA
 
 | | NMR structure of 5'-r(GGAUGCCUCCCGAGUGCAUCC): an RNA hairpin derived from the mouse 5'ETS that binds nucleolin RBD12. | | Descriptor: | 18S ribosomal RNA, 5'ETS | | Authors: | Finger, L.D, Trantirek, L, Johansson, C, Feigon, J. | | Deposit date: | 2003-09-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of Stem-loop RNAs that Bind to the Two N-terminal RNA Binding Domains of Nucleolin
Nucleic Acids Res., 31, 2003
|
|
1U6J
 
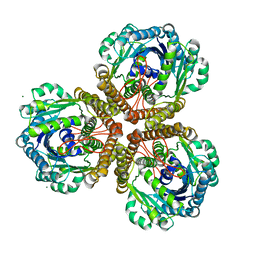 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.4A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Q3G
 
 | | MurA (Asp305Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, 3'-1-CARBOXY-1-PHOSPHONOOXY-ETHOXY-URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
1PPZ
 
 | | Trypsin complexes at atomic and ultra-high resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PJL
 
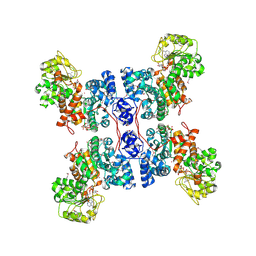 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
1PQ8
 
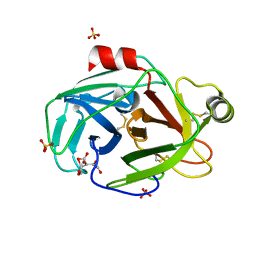 | | Trypsin at pH 4 at atomic resolution | | Descriptor: | CITRIC ACID, GLY-GLY-ARG PEPTIDE, LYSINE, ... | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1OII
 
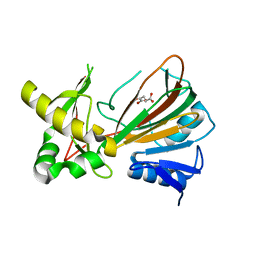 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with iron and alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OJ7
 
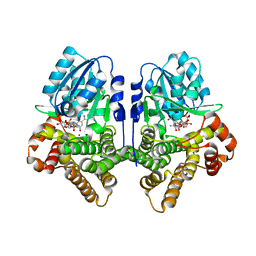 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
1RFU
 
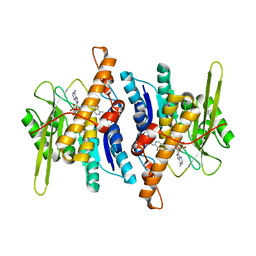 | | Crystal structure of pyridoxal kinase complexed with ADP and PLP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, ZINC ION, ... | | Authors: | Liang, D.-C, Jiang, T, Li, M.-H. | | Deposit date: | 2003-11-10 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes in the reaction of pyridoxal kinase
J.BIOL.CHEM., 279, 2004
|
|
1RXS
 
 | | E. coli uridine phosphorylase: 2'-deoxyuridine phosphate complex | | Descriptor: | 2'-DEOXYURIDINE, META VANADATE, PHOSPHATE ION, ... | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
1S67
 
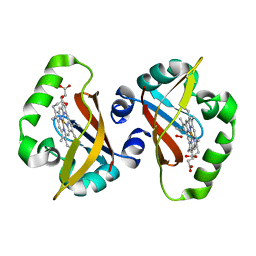 | | Crystal structure of heme domain of direct oxygen sensor from E. coli | | Descriptor: | Hypothetical protein yddU, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, H.J, Suquet, C, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH)
Biochemistry, 43, 2004
|
|
1Q1F
 
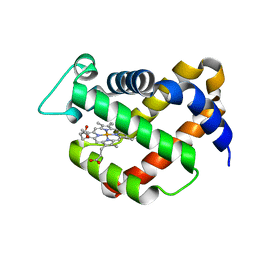 | | Crystal structure of murine neuroglobin | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vallone, B, Nienhaus, K, Matthes, K, Brunori, M, Nienhaus, G.U. | | Deposit date: | 2003-07-19 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of murine neuroglobin: Novel pathways for ligand migration and binding.
Proteins, 56, 2004
|
|
1PQ3
 
 | | Human Arginase II: Crystal Structure and Physiological Role in Male and Female Sexual Arousal | | Descriptor: | Arginase II, mitochondrial precursor, CHLORIDE ION, ... | | Authors: | Cama, E, Colleluori, D.M, Emig, F.A, Shin, H, Kim, S.W, Kim, N.N, Traish, A.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-06-17 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Arginase II: Crystal Structure and Physiological Role in Male and Female Sexual Arousal
Biochemistry, 42, 2003
|
|
