5N37
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule N-(1,3-benzodioxol-5-ylmethyl)cyclopentanamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-benzodioxol-5-ylmethyl(cyclopentyl)azanium, DIMETHYL SULFOXIDE, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | A crystallographic fragment study with cAMP-dependent protein kinase A
To Be Published
|
|
7CHQ
 
 | | AcrIE2 | | Descriptor: | anti-CRISPR AcrIE2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A 1.3 angstrom high-resolution crystal structure of an anti-CRISPR protein, AcrI E2.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
6IE4
 
 | |
6U5Z
 
 | |
9OOY
 
 | |
8XG4
 
 | |
9OMC
 
 | | Crystal structure of E. coli ApaH in complex with Gp4G | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Bis(5'-nucleosyl)-tetraphosphatase [symmetrical], ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2025-05-13 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | ApaH decaps Np 4 N-capped RNAs in two alternative orientations.
Nat.Chem.Biol., 2025
|
|
8J2V
 
 | |
5MOF
 
 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form in complex with manganese and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, CHLORIDE ION, ... | | Authors: | McDonough, M.A, Zhang, Z, Schofield, C.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y2J
 
 | |
7C9J
 
 | | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Protein-glutamine gamma-glutamyltransferase | | Authors: | Takita, T, Mikami, B, Lei, Y, Jing, Y, Yamada, A, Yasukawa, K. | | Deposit date: | 2020-06-06 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension)
To be published
|
|
8PP3
 
 | |
6QGQ
 
 | | Crystal structure of APT1 C2S mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, GLYCEROL, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
7Q06
 
 | | Crystal structure of TPADO in complex with 2-OH-TPA | | Descriptor: | 2-Hydroxyterephthalic acid, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8S6R
 
 | | RosC-8.demethyl-8-amino-riboflavin complex | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, GLYCEROL, Phosphoglycerate mutase | | Authors: | Ermler, U, Warkentin, E, Demmer, U, Mack, M. | | Deposit date: | 2024-02-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Phosphatase RosC from Streptomyces davaonensis is Used for Roseoflavin Biosynthesis and has Evolved to Largely Prevent Dephosphorylation of the Important Cofactor Riboflavin-5'-phosphate.
J.Mol.Biol., 436, 2024
|
|
5NK5
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1m | | Descriptor: | 2-[[3-[(3-aminophenyl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6S1F
 
 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[3-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-5-methoxy-phenyl]methanesulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
To be published
|
|
6HJS
 
 | | Crystal structure of glutathione transferase Omega 1C from Trametes versicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 2021
|
|
5LLT
 
 | | Plasmodium falciparum nicotinic acid mononucleotide adenylyltransferase complexed with NaAD | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINIC ACID ADENINE DINUCLEOTIDE, ... | | Authors: | Bathke, J, Fritz-Wolf, K, Brandstaedter, C, Rahlfs, S, Becker, K. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Characterization of Plasmodium falciparum Nicotinic Acid Mononucleotide Adenylyltransferase.
J. Mol. Biol., 428, 2016
|
|
8K0M
 
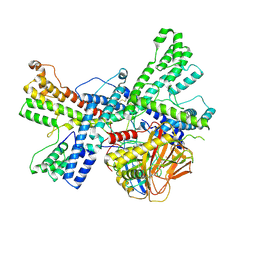 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0F
 
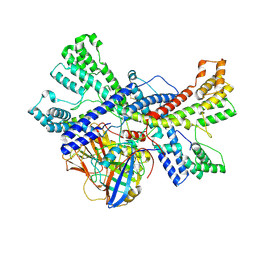 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0I
 
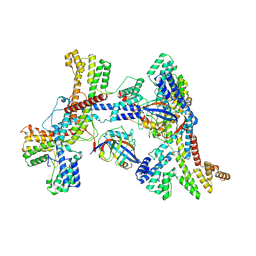 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its dual-ternary state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, Peptidyl-prolyl cis-trans isomerase B, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
9E2B
 
 | | Structure of a solute binding protein from Desulfonauticus sp. bound to L-tryptophan | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transporter, periplasmic subunit family 3, ... | | Authors: | Rahman, M, Frkic, R.L, Smith, O.B, Jackson, C.J. | | Deposit date: | 2024-10-22 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a solute binding protein from Desulfonauticus sp. bound to L-tryptophan
To Be Published
|
|
8X7G
 
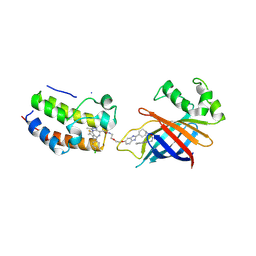 | | Crystal structure of the ternary complex of GID4-PROTAC(NEP108)-BRD4(BD1). | | Descriptor: | 2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]-~{N}-[2-[2-[[2-[4-[2-(1~{H}-indol-2-ylmethylamino)ethanoylamino]cyclohexyl]-3~{H}-benzimidazol-5-yl]oxy]ethoxy]ethyl]ethanamide, Bromodomain-containing protein 4, Glucose-induced degradation protein 4 homolog | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2023-11-24 | | Release date: | 2025-03-26 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of PROTACs utilizing the E3 ligase GID4 for targeted protein degradation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
