8EWE
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWQ
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]benzamide}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWR
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8F61
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine Quasi-Anaerobically | | Descriptor: | (5S)-5-methyl-1,3-diazinane-2,4-dione, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8EWD
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8QT1
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-5 | | Descriptor: | (2S)-2-dodecylsulfanylpropanoic acid, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
6NTL
 
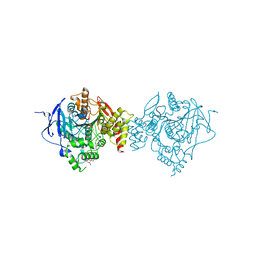 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-234 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
8RLN
 
 | | Crystal structure of human adenosine A2A receptor (construct A2A-PSB2-bRIL) complexed with the partial antagonist LUF5834 at the orthosteric pocket | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-azanyl-4-(4-hydroxyphenyl)-6-(1~{H}-imidazol-2-ylmethylsulfanyl)pyridine-3,5-dicarbonitrile, ... | | Authors: | Strater, N, Claff, T, Weisse, R.H, Muller, C.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Partial Activation of the Prototypic G Protein-Coupled Adenosine A 2A Receptor.
Acs Pharmacol Transl Sci, 7, 2024
|
|
6GFI
 
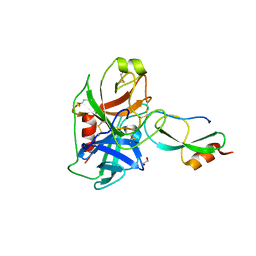 | | Structure of Human Mesotrypsin in complex with APPI variant T11V/M17R/I18F/F34V | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, PRSS3 protein | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N, Naftaly, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries.
Nat Commun, 9, 2018
|
|
4Q7G
 
 | | 1.7 Angstrom Crystal Structure of leukotoxin LukD from Staphylococcus aureus. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leucotoxin LukDv | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Shatsman, S, Kwon, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6Q3M
 
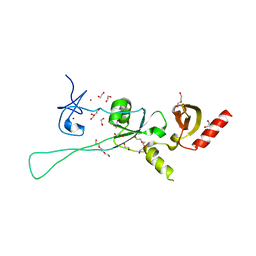 | | Structure of CHD4 PHD2 - tandem chromodomains | | Descriptor: | 1,2-ETHANEDIOL, Chromodomain-helicase-DNA-binding protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alt, A, Mancini, E.J. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of histone readers
To Be Published
|
|
9LG2
 
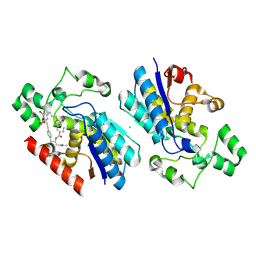 | | Phosphoglycerate mutase 1 complexed with a covalent inhibitor | | Descriptor: | CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Li, S, Wu, X, Zhou, L, Lu, X. | | Deposit date: | 2025-01-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5J44
 
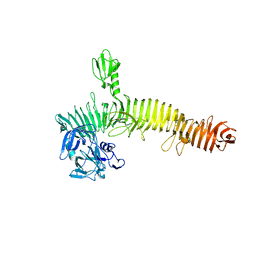 | | Crystal structure of the Secreted Extracellular protein A (SepA) from Shigella flexneri | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Serine protease SepA autotransporter | | Authors: | Birtley, J.R, Stern, L.J, McCormick, B, Maldonado-Contreras, A. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.912 Å) | | Cite: | Shigella depends on SepA to destabilize the intestinal epithelial integrity via cofilin activation.
Gut Microbes, 8, 2017
|
|
6QKV
 
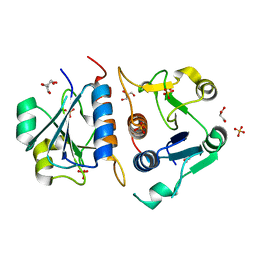 | | Structure of YibK from P. aeruginosa | | Descriptor: | GLYCEROL, SULFATE ION, tRNA (cytidine(34)-2'-O)-methyltransferase | | Authors: | Mikula, K.M, Tascon, I, Iwai, H. | | Deposit date: | 2019-01-30 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tying up the Loose Ends: A Mathematically Knotted Protein.
Front Chem, 9, 2021
|
|
8A0Z
 
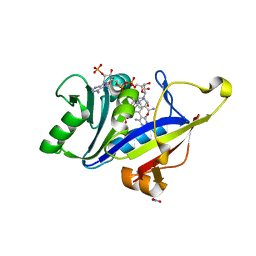 | |
5MHL
 
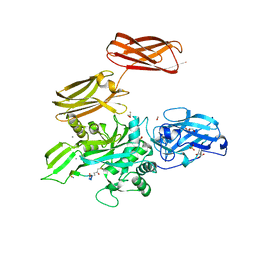 | |
8U16
 
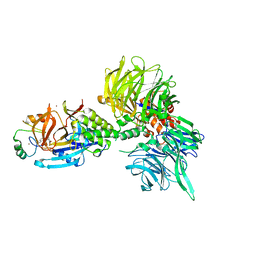 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
6QH3
 
 | |
8KC9
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to cyclosporin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, DSN-MLE-MLE-MVA-BMT-ABA-SAR-MLE-VAL-MLE-ALA, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
7MMC
 
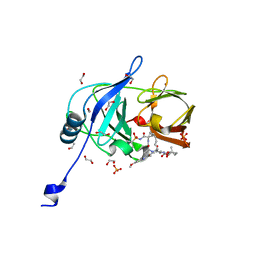 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6NBS
 
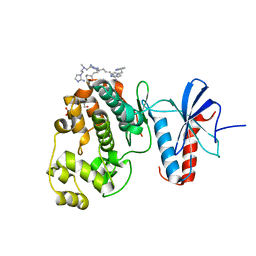 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
8P04
 
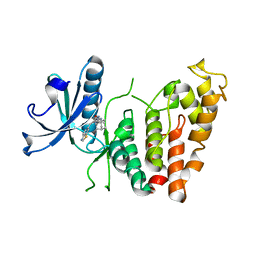 | | Crystal structure of human CLK1 in complex with Leucettinib-92 | | Descriptor: | (4~{Z})-2-(1-adamantylamino)-4-(1,3-benzothiazol-6-ylmethylidene)-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Leucettinibs, a Class of DYRK/CLK Kinase Inhibitors Inspired by the Marine Sponge Natural Product Leucettamine B.
J.Med.Chem., 66, 2023
|
|
5K4T
 
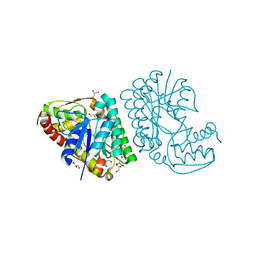 | |
8IY2
 
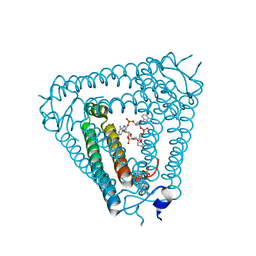 | | Structure of Acb2 complexed with 3',3'-cGAMP and cAAA | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-3'-MONOPHOSPHATE, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, ... | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8OQ7
 
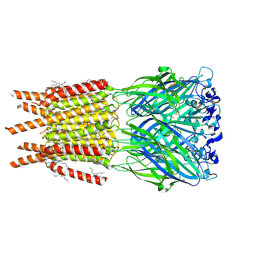 | | CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
