6SA6
 
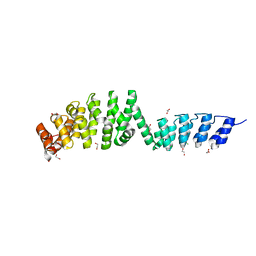 | | DARPin-Armadillo fusion A5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPin-Armadillo fusion A5 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Pluckthun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
5MVW
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5WGK
 
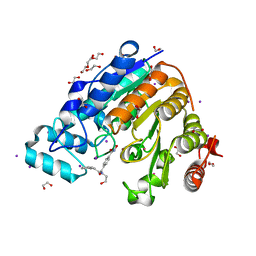 | |
4WEV
 
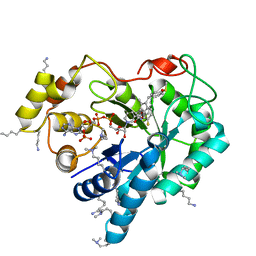 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
8Y9Q
 
 | | b-glucosidase from Thermotoga profunda Tp-BGL | | Descriptor: | GLYCEROL, b-glucosidase | | Authors: | Guo, Y, Chen, A. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights and functional characterization of a novel beta-glucosidase derived from Thermotoga profunda.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
8QRJ
 
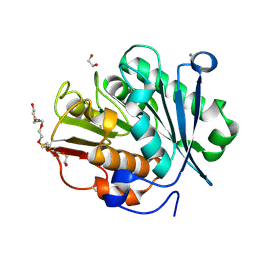 | | LCC-ICCG PETase mutant H218Y | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, ... | | Authors: | Orr, G, Niv, Y, Barakat, M, Boginya, A, Dessau, M, Afriat-Jurnou, L. | | Deposit date: | 2023-10-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Streamlined screening of extracellularly expressed PETase libraries for improved polyethylene terephthalate degradation.
Biotechnol J, 19, 2024
|
|
8ELZ
 
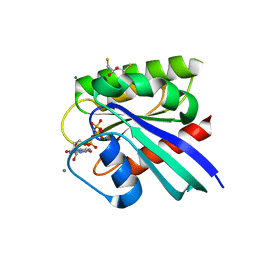 | | HRAS R97M Crystal Form 1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, GTPase HRas, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
6W5H
 
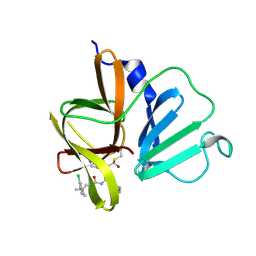 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
7M5Z
 
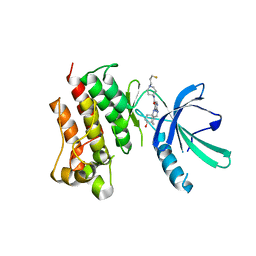 | | Crystal Structure of the MerTK Kinase Domain in Complex with Inhibitor MIPS15692 | | Descriptor: | 2-(butylamino)-N-[1-(3-fluoropropyl)piperidin-4-yl]-4-{[(1r,4r)-4-hydroxycyclohexyl]amino}pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Hermans, S.J, Hancock, N.C, Baell, J.B, Parker, M.W. | | Deposit date: | 2021-03-25 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Development of [ 18 F]MIPS15692, a radiotracer with in vitro proof-of-concept for the imaging of MER tyrosine kinase (MERTK) in neuroinflammatory disease.
Eur.J.Med.Chem., 226, 2021
|
|
7QNJ
 
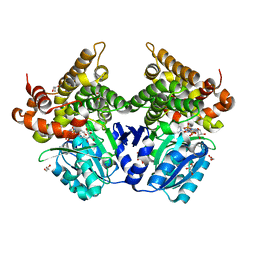 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NAD+, AND GLYCEROL | | Descriptor: | FE (III) ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
5WIM
 
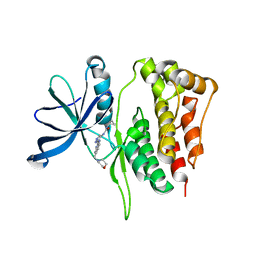 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
8KIH
 
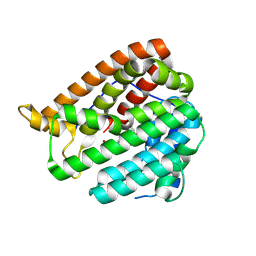 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-23 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Phomactin Platelet Activating Factor Antagonist Requires a Two-Enzyme Cascade.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6O07
 
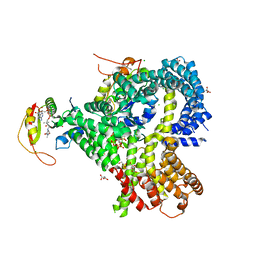 | |
8RLN
 
 | | Crystal structure of human adenosine A2A receptor (construct A2A-PSB2-bRIL) complexed with the partial antagonist LUF5834 at the orthosteric pocket | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-azanyl-4-(4-hydroxyphenyl)-6-(1~{H}-imidazol-2-ylmethylsulfanyl)pyridine-3,5-dicarbonitrile, ... | | Authors: | Strater, N, Claff, T, Weisse, R.H, Muller, C.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Partial Activation of the Prototypic G Protein-Coupled Adenosine A 2A Receptor.
Acs Pharmacol Transl Sci, 7, 2024
|
|
7OSQ
 
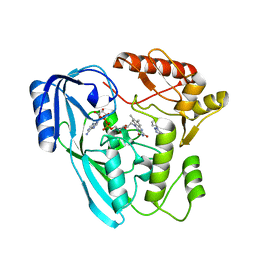 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
4X7G
 
 | | CobK precorrin-6A reductase | | Descriptor: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | Authors: | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
8T35
 
 | | Crystal structure of K51 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T36
 
 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
6O49
 
 | |
6QJU
 
 | |
6O23
 
 | | Crystal structure of 2243 Fab in complex with circumsporozoite protein NANP5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2243 Fab heavy chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6QHK
 
 | |
8K9X
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 5 (ADKI5) | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-[3-[2-[[4-[(2,5-dimethoxyphenyl)amino]-1,3,5-triazin-2-yl]amino]phenyl]sulfonylpropyl]hexanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9W
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 4 (ADKI4) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase, ~{N}2-(2-methoxyphenyl)-~{N}4-(2-propan-2-ylsulfonylphenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9U
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 2 (ADKI2) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
