1MRS
 
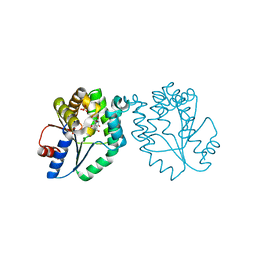 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH 5-CH2OH DEOXYURIDINE MONOPHOSPHATE | | Descriptor: | 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
1D45
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-25 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D63
 
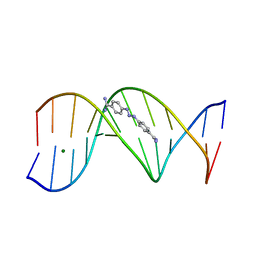 | | CRYSTAL STRUCTURE OF A BERENIL-D(CGCAAATTTGCG) COMPLEX; AN EXAMPLE OF DRUG-DNA RECOGNITION BASED ON SEQUENCE-DEPENDENT STRUCTURAL FEATURES | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Brown, D.G, Sanderson, M.R, Garman, E, Neidle, S. | | Deposit date: | 1992-03-02 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features.
J.Mol.Biol., 226, 1992
|
|
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1E2L
 
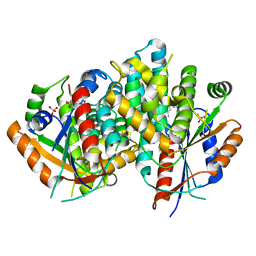 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-09-14 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
1DN8
 
 | |
1DXN
 
 | |
1E2I
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | 9-HYDROXYPROPYLADENINE, R-ISOMER, S-ISOMER, ... | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins, 41, 2000
|
|
1E2J
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
1E2H
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
1E2K
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
4ASJ
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-amino-1-benzyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-N-methylbenzenesulfonamide, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
1FKZ
 
 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
101D
 
 | |
113D
 
 | | THE STRUCTURE OF GUANOSINE-THYMIDINE MISMATCHES IN B-DNA AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Hunter, W.N, Brown, T, Kneale, G, Anand, N.N, Rabinovich, D, Kennard, O. | | Deposit date: | 1993-01-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution.
J.Biol.Chem., 262, 1987
|
|
179D
 
 | |
1A9I
 
 | |
2FDF
 
 | | Crystal Structure of AlkB in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2ES2
 
 | |
2FDG
 
 | | Crystal Structure of AlkB in complex with Fe(II), succinate, and methylated trinucleotide T-meA-T | | Descriptor: | 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, FE (II) ION, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDI
 
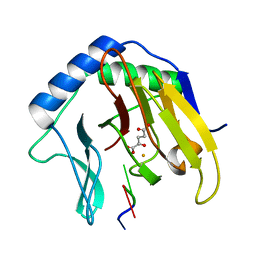 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T (air 3 hours) | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FD8
 
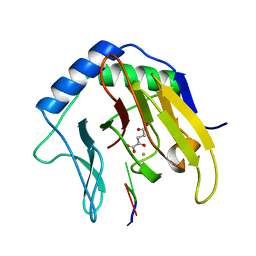 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
1A9H
 
 | |
1A9G
 
 | |
