7FUE
 
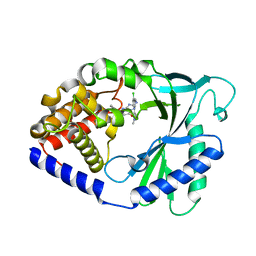 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[[2-chloro-5-(1-methylpyrazol-3-yl)phenyl]methylamino]-5-(2-phenylethyl)-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-2-({[(5P)-2-chloro-5-(1-methyl-1H-pyrazol-3-yl)phenyl]methyl}amino)-5-(2-phenylethyl)[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.169 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUJ
 
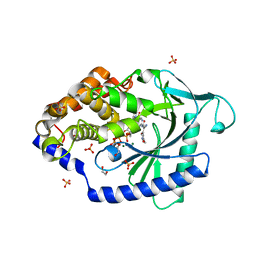 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-2-cyano-N-[4-(3-fluorophenyl)-5-methylsulfonylpyrimidin-2-yl]-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-2-cyano-N-[(4P)-4-(3-fluorophenyl)-5-(methanesulfonyl)pyrimidin-2-yl]-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUD
 
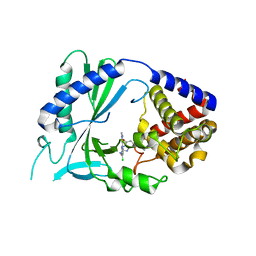 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 5-benzyl-2-[[2-chloro-5-(1-methylpyrazol-3-yl)phenyl]methylamino]-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-5-benzyl-2-({[(5P)-2-chloro-5-(1-methyl-1H-pyrazol-3-yl)phenyl]methyl}amino)[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUR
 
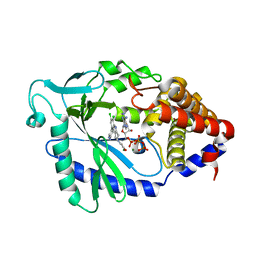 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydropyrido[4,3-b]indol-2-yl]-2-hydroxyethanone | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUP
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with methyl 1-benzoyl-3-methyl-5-(1,2-oxazol-5-yl)pyrazole-4-carboxylate | | Descriptor: | Cyclic GMP-AMP synthase, PROPANOIC ACID, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FU1
 
 | |
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VP4
 
 | |
2W7X
 
 | | Cellular inhibition of checkpoint kinase 2 and potentiation of cytotoxic drugs by novel Chk2 inhibitor PV1019 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[4-[(E)-N-carbamimidamido-C-methyl-carbonimidoyl]phenyl]-7-nitro-1H-indole-2-carboxamide, ... | | Authors: | Jobson, A.G, Lountos, G.T, Lorenzi, P.L, Llamas, J, Connelly, J, Tropea, J.E, Onda, A, Kondapaka, S, Zhang, G, Caplen, N.J, Caredellina, J.H, Monks, A, Self, C, Waugh, D.S, Shoemaker, R.H, Pommier, Y. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cellular Inhibition of Chk2 Kinase and Potentiation of Camptothecins and Radiation by the Novel Chk2 Inhibitor Pv1019.
J.Pharmacol.Exp.Ther., 331, 2009
|
|
6U5Z
 
 | |
6USM
 
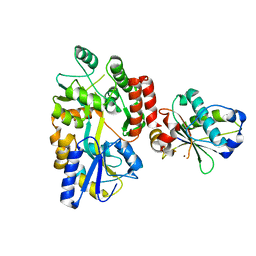 | |
8JDI
 
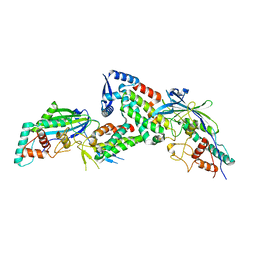 | |
8JDH
 
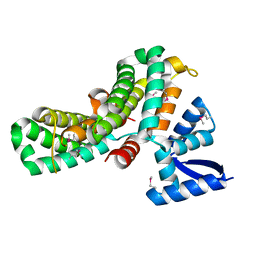 | |
4EC8
 
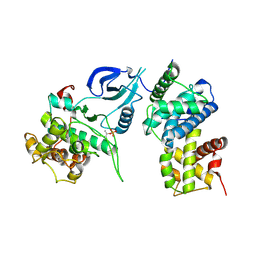 | |
6V8I
 
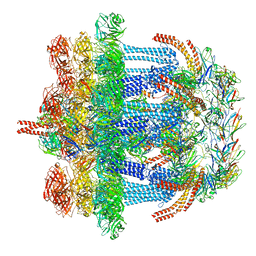 | |
7TGP
 
 | | E176T/Y188G variant of the internal UBA Domain of HHR23A | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Rothfuss, M, Bowler, B.E, McClelland, L.J, Sprang, S.R. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
6V12
 
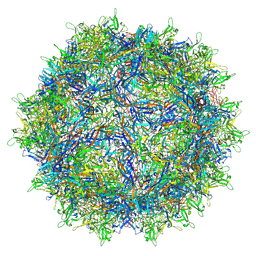 | | Empty AAV8 particles | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6UUE
 
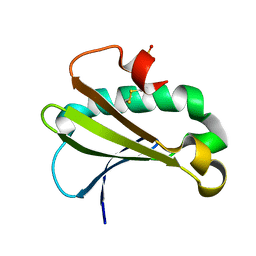 | |
8JVM
 
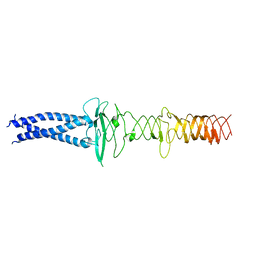 | | AHS-CSF domains of phage lambda tail | | Descriptor: | Tip attachment protein J | | Authors: | Wang, J. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8KGE
 
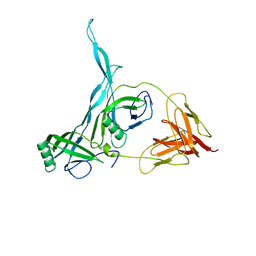 | |
8K37
 
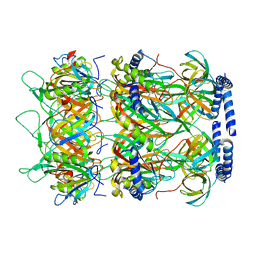 | | Structure of the bacteriophage lambda neck | | Descriptor: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K36
 
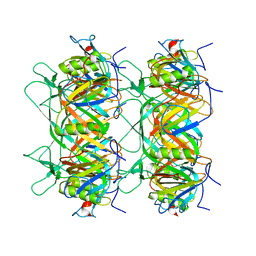 | |
8K35
 
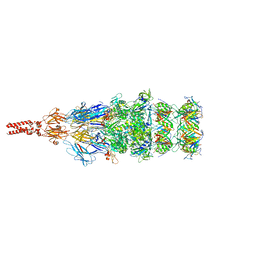 | | Structure of the bacteriophage lambda tail tip complex | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
6V1T
 
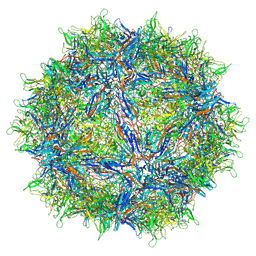 | | Empty AAVrh.39 particle | | Descriptor: | Capsid protein VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6UTC
 
 | |
