4U2J
 
 | |
8BWB
 
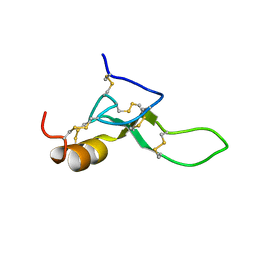 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
7MRZ
 
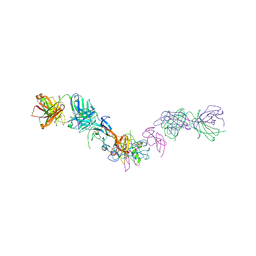 | | Structure of GDF11 bound to fused ActRIIB-ECD and Alk4-ECD with Anti-ActRIIB Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B,Activin receptor type-1B, Fab Heavy Chain, ... | | Authors: | Goebel, E.J, Kattamuri, C, Gipson, G.R, Thompson, T.B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
2BAX
 
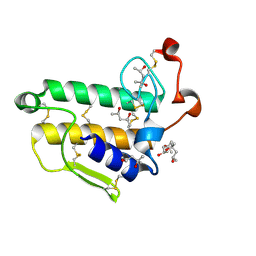 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7MEP
 
 | |
7MDU
 
 | |
7COY
 
 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
7CJI
 
 | | Photosystem II structure in the S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
9EUP
 
 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
9EUO
 
 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
4UB8
 
 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
4UB6
 
 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
8I1C
 
 | |
8I1A
 
 | |
8I1I
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1D
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I19
 
 | |
8I18
 
 | |
8I1H
 
 | |
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I8T
 
 | |
8I1F
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.6 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
