5VQP
 
 | | Crystal structure of human pro-TGF-beta1 | | Descriptor: | Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-09 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
4EG4
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1289 | | Descriptor: | 2-({3-[(3,5-dibromo-2-ethoxybenzyl)amino]propyl}amino)quinolin-4(1H)-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4A6Z
 
 | | Cytochrome c peroxidase with bound guaiacol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, Guaiacol, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Nnamchi, C, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
4EGA
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1320 | | Descriptor: | 2-({3-[(3,5-dibromo-2-methoxybenzyl)amino]propyl}amino)quinolin-4(1H)-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4FN9
 
 | | X-ray Crystal structure of the Ancestral 3-keto steroid receptor - Progesterone complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Colucci, J.K, Ortlund, E.A, Thornton, J.W. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evolution of minimal specificity and promiscuity in steroid hormone receptors.
PLoS Genet., 8, 2012
|
|
4FNE
 
 | | X-ray Crystal structure of the Ancestral 3-keto steroid receptor - DOC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESOXYCORTICOSTERONE, ... | | Authors: | Ortlund, E.A, Colucci, J.K, Thornton, J, Eick, G, Harms, M. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Evolution of minimal specificity and promiscuity in steroid hormone receptors.
PLoS Genet, 8, 2012
|
|
4B3N
 
 | | Crystal structure of rhesus TRIM5alpha PRY/SPRY domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MALTOSE-BINDING PERIPLASMIC PROTEIN, TRIPARTITE MOTIF-CONTAINING PROTEIN 5, ... | | Authors: | Yang, H, Ji, X, Zhao, Q, Xiong, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insight Into HIV-1 Capsid Recognition by Rhesus Trim5Alpha
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HPP
 
 | |
8F0H
 
 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
6TL8
 
 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | Authors: | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
5H6U
 
 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate pentasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
1C47
 
 | |
5HET
 
 | |
5HGW
 
 | |
1J8T
 
 | | Catalytic Domain of Human Phenylalanine Hydroxylase Fe(II) | | Descriptor: | FE (II) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin.
J.Mol.Biol., 314, 2001
|
|
7ZET
 
 | | Crystal structure of human Clusterin, crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin, ... | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Clusterin, crystal form I
To be published
|
|
7ZEU
 
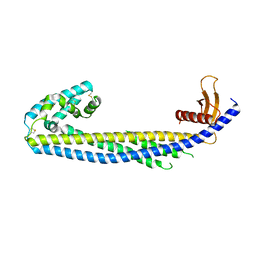 | |
1ILE
 
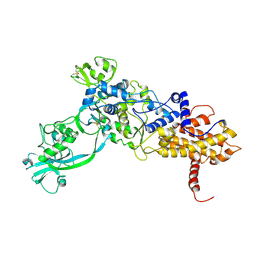 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
8A3T
 
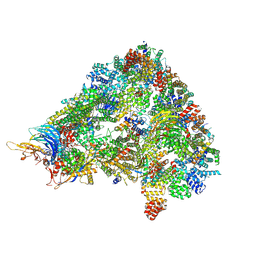 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
5IQ5
 
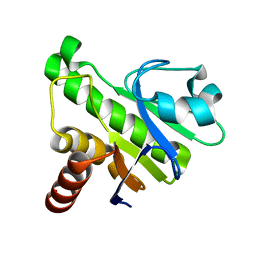 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
5I3M
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated thiourea-linked carboxylate zinc-chelator water-soluble inhibitor (DC31). | | Descriptor: | (2S)-2-{[2-({[(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]carbamothioyl}amino)ethyl](biphenyl-4-ylsulfonyl)amino}-3-methylbutanoic acid (non-preferred name), 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
5I13
 
 | | Endonuclease inhibitor 2 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
8T9A
 
 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | Authors: | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
To Be Published
|
|
7ODM
 
 | | AmGSTF1 Y118S variant | | Descriptor: | Glutathione transferase, [(2~{S})-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-sulfanyl-propan-2-yl]amino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]azanium | | Authors: | Pohl, E, Eno, R.F.M. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
1J8U
 
 | | Catalytic Domain of Human Phenylalanine Hydroxylase Fe(II) in Complex with Tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, FE (II) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin.
J.Mol.Biol., 314, 2001
|
|
