5VUA
 
 | | Pim1 Kinase in complex with a benzofuranone inhibitor | | Descriptor: | (2Z)-6-methoxy-7-(piperazin-1-ylmethyl)-2-(1H-pyrrolo[2,3-c]pyridin-3-ylmethylidene)-1-benzofuran-3-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Theoretical Analysis of Activity Cliffs among Benzofuranone-Class Pim1 Inhibitors Using the Fragment Molecular Orbital Method with Molecular Mechanics Poisson-Boltzmann Surface Area (FMO+MM-PBSA) Approach
J Chem Inf Model, 57, 2017
|
|
8JLK
 
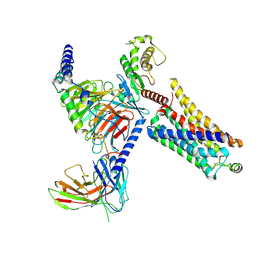 | | Ulotaront(SEP-363856)-bound mTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6BKD
 
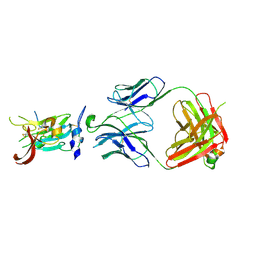 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3D heavy chain, Fab AR3D light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
5W0F
 
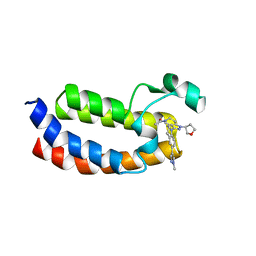 | | CREBBP Bromodomain in complex with Cpd3 ((S)-1-(3-(6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0L
 
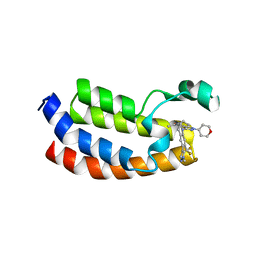 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
1GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E/R37W MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1996-12-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
4XX5
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
8CE1
 
 | | Cytochrome c maturation complex CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
4XZ4
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
9HTX
 
 | |
9DB2
 
 | | Class Ia ribonucleotide reductase with mechanism-based inhibitor N3CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{(3xi)-3-C-amino-2-deoxy-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-beta-D-threo-pentofuranosyl}pyrimidin-2(1H)-one, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Westmoreland, D.E, Drennan, C.L. | | Deposit date: | 2024-08-23 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | 2.6- angstrom resolution cryo-EM structure of a class Ia ribonucleotide reductase trapped with mechanism-based inhibitor N 3 CDP.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
9HU9
 
 | | Glycosyltransferase C from the Limosilactobacillus reuteri accessory secretion system. Complex with UDP-GlcNAc. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, GLYCEROL, ... | | Authors: | Pfalzgraf, H.E, Griffiths, R, Juge, N, Hemmings, A.M. | | Deposit date: | 2024-12-21 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for the strain-dependent UDP-sugar specificity of glycosyltransferase C from the Limosilactobacillus reuteri accessory secretion system
To Be Published
|
|
6A85
 
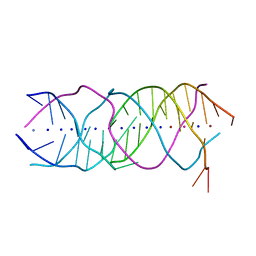 | | Crystal structure of a novel DNA quadruplex | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*GP*GP*GP*TP*GP*CP*GP*TP*T)-3'), LEAD (II) ION, ... | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2018-07-06 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads.
Nucleic Acids Res., 46, 2018
|
|
5OCX
 
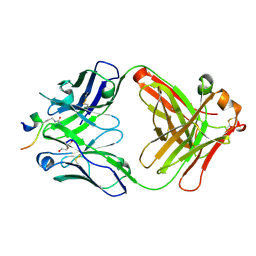 | | Crystal structure of ACPA E4 in complex with CII-C-13-CIT | | Descriptor: | 1,2-ETHANEDIOL, CII-C-13-CIT, Fab fragment anti-citrullinated protein antibody E4 - light chain, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
8OGI
 
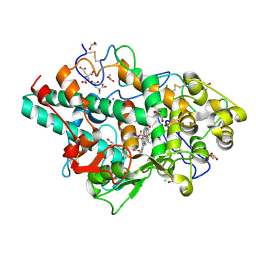 | | Structure of native human eosinophil peroxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pfanzagl, V, Obinger, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Posttranslational modification and heme cavity architecture of human eosinophil peroxidase-insights from first crystal structure and biochemical characterization.
J.Biol.Chem., 299, 2023
|
|
5LZP
 
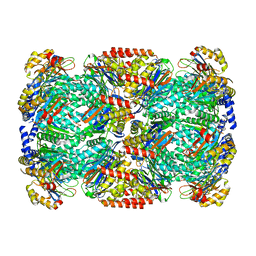 | | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | | Descriptor: | Bacterial proteasome activator, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
4Y8W
 
 | | Crystal Structure of Human Cytochrome P450 21A2 Progesterone Complex | | Descriptor: | Cytochrome P450 21-hydroxylase, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pallan, P.S, Lei, L, Egli, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Human Cytochrome P450 21A2, the Major Steroid 21-Hydroxylase: STRUCTURE OF THE ENZYMEPROGESTERONE SUBSTRATE COMPLEX AND RATE-LIMITING C-H BOND CLEAVAGE.
J.Biol.Chem., 290, 2015
|
|
8G35
 
 | | Crystal structure of F182L-CYP199A4 in complex with (S)-4-(2-hydroxy-3-oxobutan-2-yl)benzoic acid | | Descriptor: | 4-[(2S)-2-hydroxy-3-oxobutan-2-yl]benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bell, S.G, Bruning, J.B. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
8G36
 
 | | Crystal structure of F182L-CYP199A4 in complex with terephthalic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
1A6B
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | Authors: | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | Deposit date: | 1998-02-23 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
4X4D
 
 | |
8JLO
 
 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
