5OD9
 
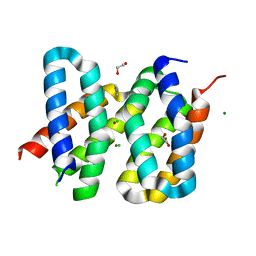 | | Structure of the engineered metalloesterase MID1sc9 | | Descriptor: | (2~{S})-2-phenylpropanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Studer, S, Mittl, P.R.E, Hilvert, D. | | Deposit date: | 2017-07-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Evolution of a highly active and enantiospecific metalloenzyme from short peptides.
Science, 362, 2018
|
|
5OP3
 
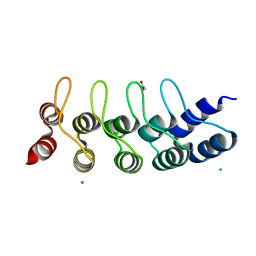 | | Designed Ankyrin Repeat Protein (DARPin) NDNH-1 selected by directed evolution against Lysozyme | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fischer, G, Hogan, B.J, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.359 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) NDNH-1 selected by directed evolution against Lysozyme
To be published
|
|
7Y7P
 
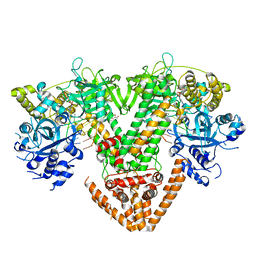 | | QDE-1 in complex with RNA template, RNA primer and AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7Q
 
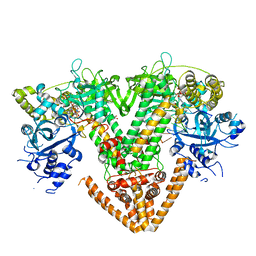 | | QDE-1 in complex with RNA template, RNA primer and 3'-dGTP | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7R
 
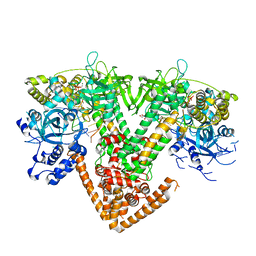 | | QDE-1 in complex with DNA template, RNA primer and 3'-dGTP | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*AP*AP*CP*TP*AP*CP*CP*GP*TP*CP*GP*GP*A)-3'), ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7S
 
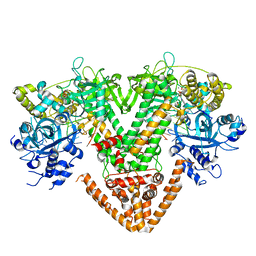 | | QDE-1 in complex with DNA template, RNA primer and AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7T
 
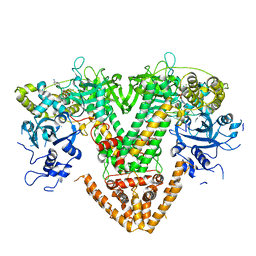 | | QDE-1 in complex with 12nt DNA template, ATP and 3'-dGTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*G*AP*GP*AP*CP*CP*TP*TP*TP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Z7C
 
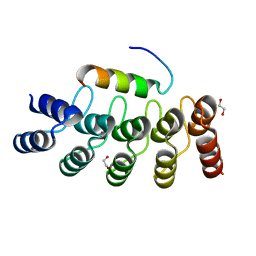 | |
7ZBS
 
 | |
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
7ZYU
 
 | |
2VUX
 
 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
5KWD
 
 | |
2W6Z
 
 | |
2W6O
 
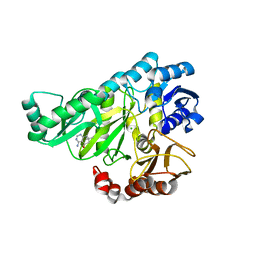 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W9J
 
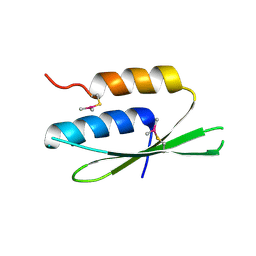 | | The crystal structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle | | Descriptor: | SIGNAL RECOGNITION PARTICLE SUBUNIT SRP14 | | Authors: | Brooks, M.A, Ravelli, R.B.G, McCarthy, A.A, Strub, K, Cusack, S. | | Deposit date: | 2009-01-25 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Srp14 from the Schizosaccharomyces Pombe Signal Recognition Particle.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5L0P
 
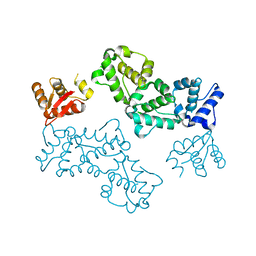 | |
5LE2
 
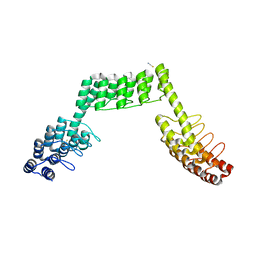 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_15_D12_15_D12 | | Descriptor: | ACETATE ION, DDD_D12_15_D12_15_D12, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEC
 
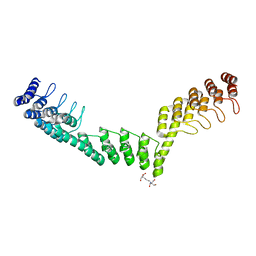 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LE9
 
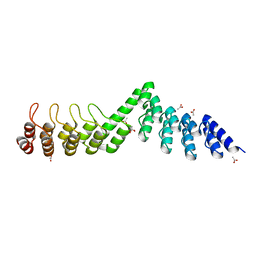 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_09_3G124 | | Descriptor: | ACETATE ION, DD_Off7_09_3G124 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEE
 
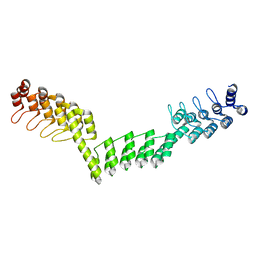 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LE8
 
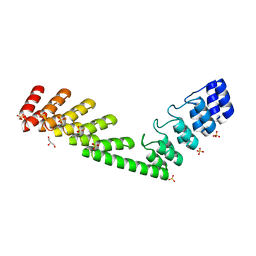 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_15_D12 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DD_D12_15_D12, GLYCEROL, ... | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LW2
 
 | | Crystal structure of DARPin 5m3_D12 | | Descriptor: | ACETATE ION, DARPin_5m3_D12 | | Authors: | Batyuk, A, Wu, Y, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LE7
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_13_D12 | | Descriptor: | DD_D12_13_D12, SULFATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LE3
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_09_D12 | | Descriptor: | DD_D12_09_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
