7JZX
 
 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF7 | | Descriptor: | AcrF7, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
5WJB
 
 | | Crystal Structure of Amino Acids 1733-1797 of Human Beta Cardiac Myosin Fused to Gp7 | | Descriptor: | Capsid assembly scaffolding protein,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
7AJU
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
2PIJ
 
 | | Structure of the Cro protein from prophage Pfl 6 in Pseudomonas fluorescens Pf-5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, Prophage Pfl 6 Cro, ... | | Authors: | Roessler, C.G, Roberts, S.A, Montfort, W.R, Cordes, M.H.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6XZP
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 4 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
3KFC
 
 | | Complex Structure of LXR with an agonist | | Descriptor: | 4-{3-[3-(methylsulfonyl)phenoxy]phenyl}-8-(trifluoromethyl)quinoline, Oxysterols receptor LXR-beta | | Authors: | Olland, A, Bernotas, R.C, Unwalla, R. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-(3-Aryloxyaryl)quinoline sulfones are potent liver X receptor agonists.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7AJB
 
 | | bovine ATP synthase dimer state1:state1 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T1J
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
7JZW
 
 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF4 | | Descriptor: | CRISPR repeat sequence, CRISPR type I-F/YPEST-associated protein Csy1, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
6YIK
 
 | | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand | | Descriptor: | (3~{R})-~{N}-[3-(3,4-dihydro-2~{H}-quinolin-1-yl)-2,2-bis(fluoranyl)propyl]-3-methyl-2-oxidanylidene-3,4-dihydro-1~{H}-quinoxaline-5-carboxamide, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand
To Be Published
|
|
4X7Q
 
 | | PIM2 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-2 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3DV8
 
 | |
7AJE
 
 | | bovine ATP synthase dimer state2:state1 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5E5B
 
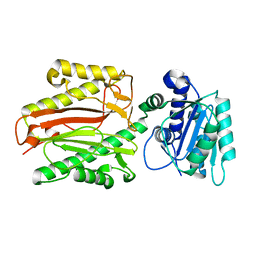 | |
6YB4
 
 | |
7AJI
 
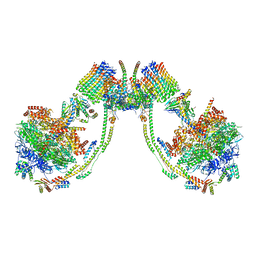 | | bovine ATP synthase dimer state3:state2 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7UGE
 
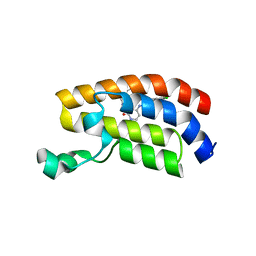 | | Bromodomain of CBP liganded with BMS-536924 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 1,2-ETHANEDIOL, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
7UGI
 
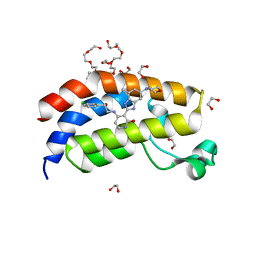 | | Bromodomain of EP300 liganded with BMS-536924 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
7AJG
 
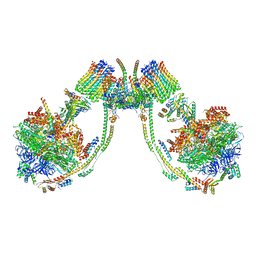 | | bovine ATP synthase dimer state2:state3 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7UGL
 
 | | Bromodomain of CBP liganded with BMS-536924 and SGC-CBP30 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
7JZZ
 
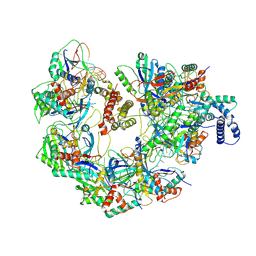 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF14 | | Descriptor: | AcrF14, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
6S57
 
 | |
2OGG
 
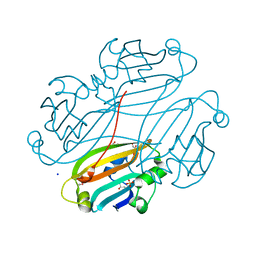 | | Structure of B. subtilis trehalose repressor (TreR) effector binding domain | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Rezacova, P, Krejcirikova, V, Borek, D, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly.
Proteins, 69, 2007
|
|
3X31
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 7,8-cis-14-epi-1a,25-Dihydroxy-19-norvitamin D3 | | Descriptor: | (1R,3R,7Z,14beta,17alpha)-17-[(2R)-6-hydroxy-6-methylheptan-2-yl]-9,10-secoestra-5,7-diene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Revisiting the 7,8-cis-vitamin D3 derivatives: synthesis, evaluating the biological activity, and study of the binding configuration
To be Published
|
|
4ZP8
 
 | | Coxsackievirus B3 Polymerase - F364L mutant | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
