7TN7
 
 | |
5KF7
 
 | |
6K3Y
 
 | | G-quadruplex complex with cyclic dinucleotide 3'-3' cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, DNA (5'-D(*TP*TP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | Authors: | Winnerdy, F.R, Heddi, B, Phan, A.T. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a G-Quadruplex Bound to Linear- and Cyclic-Dinucleotides.
J.Am.Chem.Soc., 141, 2019
|
|
6TLL
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5,6,7-TETRABROMOBENZOTRIAZOLE (tBBT) | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
8BA6
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
6X4O
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11-dimethyl-14-(propan-2-yl)-3-oxa-9,12,15,21,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(26),18,20,22,24,27-hexaene-4,10,13,16-tetrone (compound 21) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11-dimethyl-14-(propan-2-yl)-3-oxa-9,12,15,21,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(26),18,20,22,24,27-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
2XN2
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
6QSS
 
 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
8BDK
 
 | |
8OQ7
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
7RAN
 
 | | 5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (3R)-3-methyl-5-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,3,6-tetrahydropyridin-1-ium, 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Bespoke library docking for 5-HT 2A receptor agonists with antidepressant activity.
Nature, 610, 2022
|
|
5HZ5
 
 | |
8SUA
 
 | | Structure of M. baixiangningiae DarR-ligand complex | | Descriptor: | 3-azanyl-3-(hydroxymethyl)-1,5,7,11-tetraoxa-6$l^{4}-boraspiro[5.5]undecan-9-ol, DarR | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
6WO8
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5-PP-IP4), Mg, and Fluoride ion | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
7RMJ
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor (S)-N-(1-(3-(4-chloro-3-(methylsulfonamido)-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl)-6-(3-methyl-3-(methylsulfonyl)but-1-yn-1-yl)pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CAPSID PROTEIN P24, CHLORIDE ION, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and Mechanistic Bases of Viral Resistance to HIV-1 Capsid Inhibitor Lenacapavir.
Mbio, 13, 2022
|
|
5KYJ
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | (6~{R})-5-(5-fluoranyl-2-methoxy-pyrimidin-4-yl)-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazole, Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5L09
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
7Z57
 
 | | Crystal structure of Human Serum Albumin in complex with surfactant GenX (2,3,3,3-tetrafluoro-2-(heptafluoropropoxy) propanoate) | | Descriptor: | (2R)-2,3,3,3-tetrakis(fluoranyl)-2-[1,1,2,2,3,3,3-heptakis(fluoranyl)propoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, ... | | Authors: | Liberi, S, Moro, G, Vascon, F, Linciano, S, De Toni, L, Angelin, A, Cendron, L. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigation of the Interaction between Human Serum Albumin and Branched Short-Chain Perfluoroalkyl Compounds.
Chem.Res.Toxicol., 35, 2022
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8TCX
 
 | | Structure of PYCR1 complexed with 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
6XYY
 
 | | Update of ACHE FROM DROSOPHILA MELANOGASTER COMPLEX WITH TACRINE DERIVATIVE 9-(3-PHENYLMETHYLAMINO)-1,2,3,4-TETRAHYDROACRIDINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-(3-PHENYLMETHYLAMINO)-1,2,3,4-TETRAHYDROACRIDINE, Acetylcholinesterase, ... | | Authors: | Nachon, F, Sussman, J.L. | | Deposit date: | 2020-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Second Look at the Crystal Structures ofDrosophila melanogasterAcetylcholinesterase in Complex with Tacrine Derivatives Provides Insights Concerning Catalytic Intermediates and the Design of Specific Insecticides.
Molecules, 25, 2020
|
|
8TCY
 
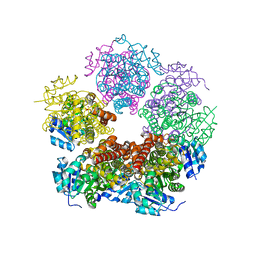 | | Structure of PYCR1 complexed with 7-fluoro-2-oxo-1,2,3,4-tetrahydroquinoline-6-carboxylic acid | | Descriptor: | 7-fluoro-2-oxo-1,2,3,4-tetrahydroquinoline-6-carboxylic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
6MDU
 
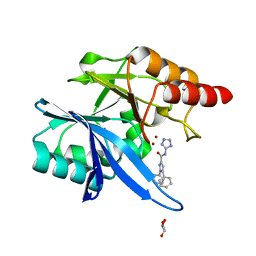 | | Crystal structure of NDM-1 with compound 7 | | Descriptor: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
1EXJ
 
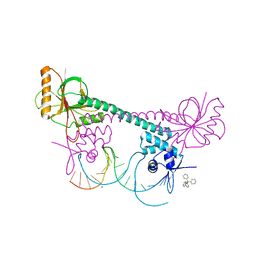 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPP | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, SODIUM ION, ... | | Authors: | Zheleznova-Heldwein, E.E, Brennan, R.G. | | Deposit date: | 2000-05-02 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
7RWQ
 
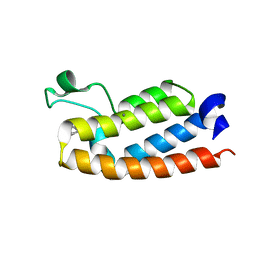 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
