3EHX
 
 | |
4QTY
 
 | | Caspase-3 E190A | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, AZIDE ION, Caspase-3, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4QU5
 
 | | Caspase-3 T140V | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4QUI
 
 | | Caspase-3 F128AV266H | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CHLORIDE ION, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4QUA
 
 | | Caspase-3 Y195F | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4QUB
 
 | | Caspase-3 K137A | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, AZIDE ION, Caspase-3 | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
3EHY
 
 | |
3Q5Y
 
 | | V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Q, Zhang, H, Wang, J.-H. | | Deposit date: | 2010-12-30 | | Release date: | 2011-03-09 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A conserved hydrophobic patch on Vbeta domains revealed by TCRbeta chain crystal structures: implications for pre-TCR dimerization
Front Immunol, 2, 2011
|
|
3QO4
 
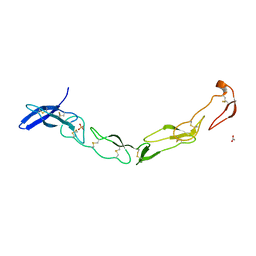 | | The Crystal Structure of Death Receptor 6 | | Descriptor: | ACETATE ION, SULFATE ION, Tumor necrosis factor receptor superfamily member 21 | | Authors: | Kuester, M, Kemmerzehl, S, Dahms, S.O, Roeser, D, Than, M.E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
J.Mol.Biol., 409, 2011
|
|
4PO4
 
 | |
3GJT
 
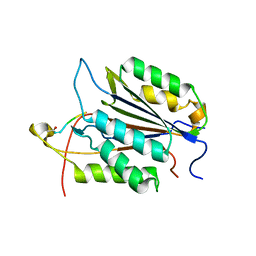 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3Q5T
 
 | |
1Q3X
 
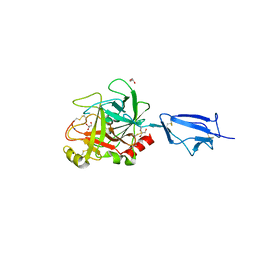 | | Crystal structure of the catalytic region of human MASP-2 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 2, SODIUM ION | | Authors: | Harmat, V, Gal, P, Kardos, J, Szilagyi, K, Ambrus, G, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sets of enzyme-substrate interactions
J.Mol.Biol., 342, 2004
|
|
1PMC
 
 | |
1A4L
 
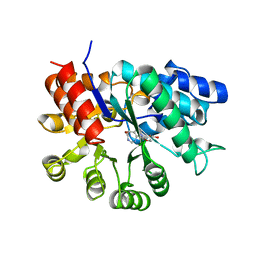 | | ADA STRUCTURE COMPLEXED WITH DEOXYCOFORMYCIN AT PH 7.0 | | Descriptor: | 2'-DEOXYCOFORMYCIN, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wang, Z, Quiocho, F.A. | | Deposit date: | 1998-01-31 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
Biochemistry, 37, 1998
|
|
3H0E
 
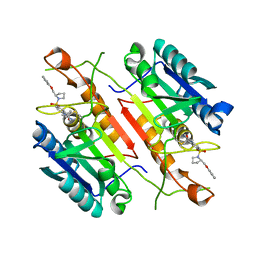 | | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as Potent Non-Peptidic Inhibitors of Caspase-3 | | Descriptor: | (10S)-3,3-dimethyl-8-{[(2S)-2-(phenoxymethyl)pyrrolidin-1-yl]sulfonyl}-2,3,4,10-tetrahydropyrimido[1,2-a]indol-10-ol, Caspase-3 | | Authors: | Xu, W. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as potent non-peptidic inhibitors of caspase-3
Bioorg.Med.Chem., 17, 2009
|
|
3R6V
 
 | | Crystal structure of aspartase from Bacillus sp. YM55-1 with bound L-aspartate | | Descriptor: | ASPARTIC ACID, Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
1A4M
 
 | | ADA STRUCTURE COMPLEXED WITH PURINE RIBOSIDE AT PH 7.0 | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wang, Z, Quiocho, F.A. | | Deposit date: | 1998-01-31 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
Biochemistry, 37, 1998
|
|
4QLU
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
1ABJ
 
 | |
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3F1A
 
 | |
4QLV
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 17 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
3F19
 
 | |
3H6N
 
 | | Crystal Structure of the ubiquitin-like domain of plexin D1 | | Descriptor: | ARSENIC, Plexin-D1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Nedyalkova, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Buck, M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of the ubiquitin-like domain of plexin D1
To be Published
|
|
