7K9S
 
 | | Cryptococcus neoformans Hsp90 nucleotide binding domain in complex with NVP-AUY922 | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, Hsp90-like protein | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Fungal-Selective Resorcylate Aminopyrazole Hsp90 Inhibitors: Optimization of Whole-Cell Anticryptococcal Activity and Insights into the Structural Origins of Cryptococcal Selectivity.
J.Med.Chem., 64, 2021
|
|
7KIV
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-10-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7KUQ
 
 | |
7KEP
 
 | | avibactam-CDD-1 2 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
7KX3
 
 | | SpeG Spermidine N-acetyltransferase F149G mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KKM
 
 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KUR
 
 | |
7KX2
 
 | | Spermidine N-acetyltransferase SpeG F149A mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KJE
 
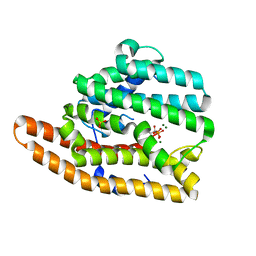 | | F96S epi-isozizaene synthase: complex with 3 Mg2+ and neridronate | | Descriptor: | (6-azanyl-1-oxidanyl-1-phosphono-hexyl)phosphonic acid, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
7KJT
 
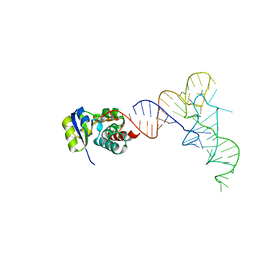 | | KEOPS tRNA modifying sub-complex of archaeal Cgi121 and tRNA | | Descriptor: | RNA (70-MER), Regulatory protein Cgi121 | | Authors: | Ceccarelli, D.F, Beenstock, J, Mao, D.Y.L, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
7KEM
 
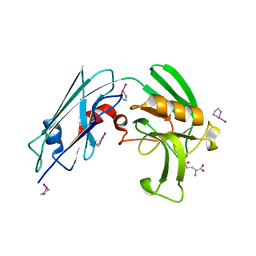 | |
7KJB
 
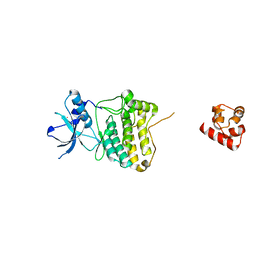 | |
7KUH
 
 | | MicroED structure of mVDAC | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Martynowycz, M.W, Khan, F, Hattne, J, Abramson, J, Gonen, T. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.12 Å) | | Cite: | MicroED structure of lipid-embedded mammalian mitochondrial voltage-dependent anion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7KUT
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N-Acetylputrescine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-aminobutyl)amino]ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7KKN
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Poly [ADP-ribose] polymerase, ... | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KWH
 
 | | Spermidine N-acetyltransferase SpeG K23-Y30 chimera from Vibrio cholerae and hSSAT | | Descriptor: | PHOSPHATE ION, Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KK3
 
 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KGV
 
 | | Crystal structure of sodium-coupled neutral amino acid transporter SLC38A9 in the N-terminal plugged form | | Descriptor: | Monoclonal antibody Fab heavy chain, Monoclonal antibody Fab light chain, Sodium-coupled neutral amino acid transporter 9 | | Authors: | Lei, H, Mu, X, Hattne, J, Gonen, T. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conformational change in the N terminus of SLC38A9 signals mTORC1 activation.
Structure, 29, 2021
|
|
7KRK
 
 | |
7KXF
 
 | |
7KUV
 
 | |
7KVL
 
 | | SARS-CoV-2 Main protease immature form - FMAX Library E01 fragment | | Descriptor: | 2-chloropyridine-4-carboxamide, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KJ8
 
 | | Wild-type epi-isozizaene synthase: complex with 3 Mg2+ and pamidronate | | Descriptor: | MAGNESIUM ION, PAMIDRONATE, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
7KJA
 
 | |
7KK4
 
 | | Structure of the catalytic domain of PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
