1TZY
 
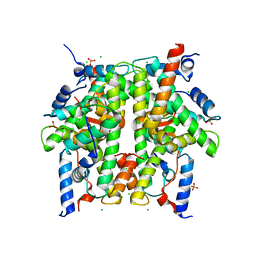 | | Crystal Structure of the Core-Histone Octamer to 1.90 Angstrom Resolution | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Nicholson, J.M, Chantalat, L, Reynolds, C.D, Lambert, S.J, Baldwin, J.P. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of the native histone octamer.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3O3L
 
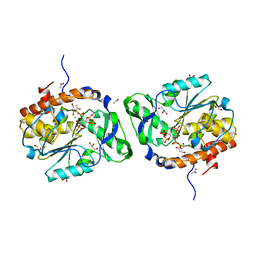 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,3,4,5)tetrakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of substrate binding in PTPLPs
To be Published
|
|
8A5Y
 
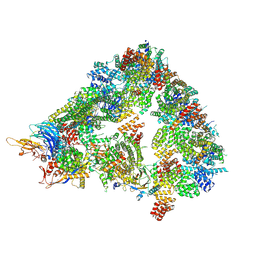 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
5AOR
 
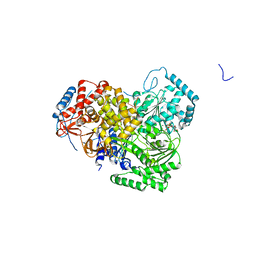 | | Structure of MLE RNA ADP AlF4 complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, DOSAGE COMPENSATION REGULATOR, ... | | Authors: | Prabu, J.R, Conti, E. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the RNA Helicase Mle Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling.
Mol.Cell, 60, 2015
|
|
8T2R
 
 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2T
 
 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8A61
 
 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
8A3T
 
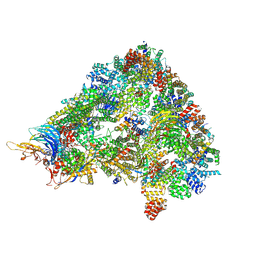 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
6HJ2
 
 | |
8TES
 
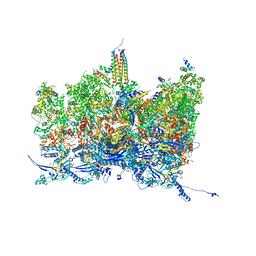 | | Human cytomegalovirus portal vertex, virion configuration 2 (VC2) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TET
 
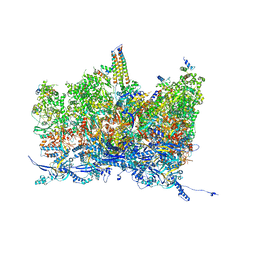 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 1 (NC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEP
 
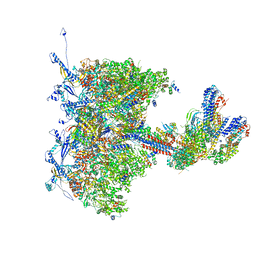 | | Human cytomegalovirus portal vertex, virion configuration 1 (VC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Inner tegument protein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEU
 
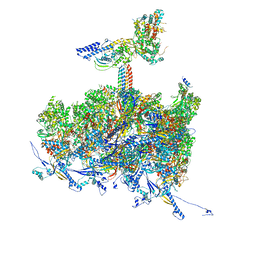 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 2 - inverted (NC2-inv) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
6R7A
 
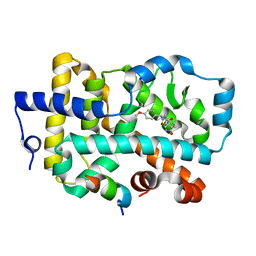 | | Ligand complex of RORg LBD | | Descriptor: | LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Nuclear receptor ROR-gamma, SODIUM ION, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Inverse Agonists of the Retinoic Acid Receptor-Related Orphan Receptor C2.
Acs Med.Chem.Lett., 10, 2019
|
|
7ZTD
 
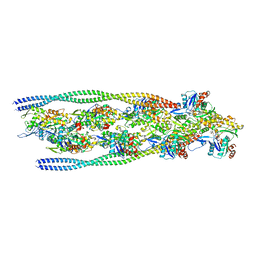 | | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 3.2, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
7ZTC
 
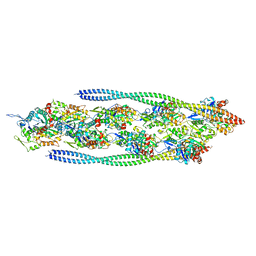 | | Non-muscle F-actin decorated with non-muscle tropomyosin 1.6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 1.6, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
6R7N
 
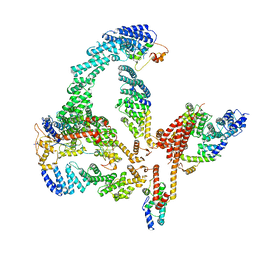 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
6HDN
 
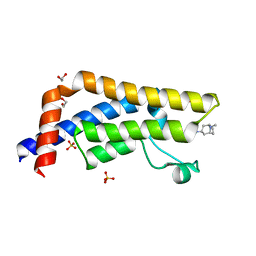 | |
7VY7
 
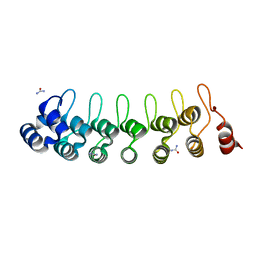 | |
7VY4
 
 | |
6S41
 
 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH XPC-7455 | | Descriptor: | 4-[[(1~{S})-1-[2,5-bis(fluoranyl)phenyl]ethyl]amino]-5-chloranyl-2-fluoranyl-~{N}-(1,3-thiazol-4-yl)benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Focken, T, Maskos, K, Griessner, A, Krapp, S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of CNS-Penetrant Aryl Sulfonamides as Isoform-Selective NaV1.6 Inhibitors with Efficacy in Mouse Models of Epilepsy.
J.Med.Chem., 62, 2019
|
|
6HN6
 
 | | A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor RXR-ALPHA | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Retinoic acid receptor RXR-alpha | | Authors: | Eberhardt, J, McEwen, A.G, Bourguet, W, Moras, D, Dejaegere, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor retinoic X receptor alpha.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6X6C
 
 | | Cryo-EM structure of NLRP1-DPP9-VbP complex | | Descriptor: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | Deposit date: | 2020-05-27 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
