9GL9
 
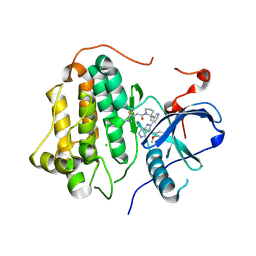 | | Wild-type EGFR bound with STX-721 | | Descriptor: | (2R,3S)-3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[2-[(2R)-1-[(E)-4-(dimethylamino)but-2-enoyl]-2-methyl-pyrrolidin-2-yl]ethynyl]pyridin-4-yl]-1,2,3,5,6,7-hexahydropyrrolo[3,2-c]pyridin-4-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 2025
|
|
6GM7
 
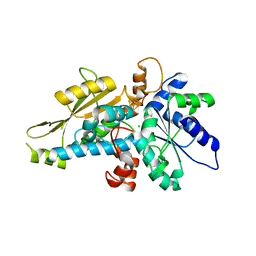 | | [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii,variant E144A | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER | | Authors: | Duan, J, Engelbrecht, V, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
8T87
 
 | |
8T88
 
 | |
6GHP
 
 | | 14-3-3sigma in complex with a TASK3 peptide stabilized by semi-synthetic natural product FC-NAc | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Potassium channel subfamily K member 9, ... | | Authors: | Andrei, S.A, de Vink, P.J, Brunsveld, L, Ottmann, C, Higuchi, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | Descriptor: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUN
 
 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
8BU4
 
 | | Structure of DDB1 bound to DS22-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUR
 
 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUF
 
 | | Structure of DDB1 bound to Z12-engaged CDK12-cyclin K | | Descriptor: | 2-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylmethyl)-6,7-dimethoxy-3~{H}-quinazolin-4-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU1
 
 | | Structure of DDB1 bound to DS17-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
1ALK
 
 | |
7AF8
 
 | | Bacterial 30S ribosomal subunit assembly complex state E (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8FVU
 
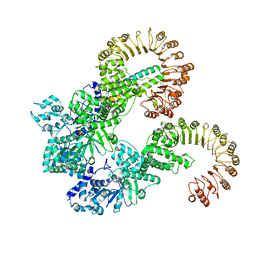 | | Cryo-EM structure of human Needle/NAIP/NLRC4 (R288A) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1, Lethal factor,Type III secretion system protein, ... | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6MWR
 
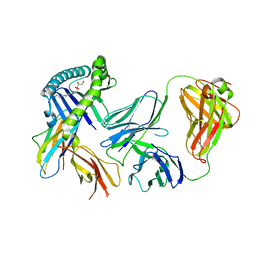 | | Recognition of MHC-like molecule | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2018-10-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Science, 366, 2019
|
|
3F84
 
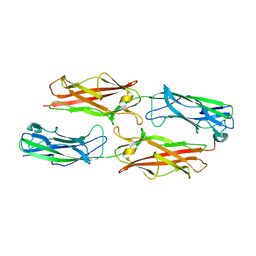 | |
7Q4K
 
 | | Erythromycin-stalled Escherichia coli 70S ribosome with streptococcal MsrDL nascent chain | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fostier, C.R, Ousalem, F, Soufari, H, Leroy, E.C, Ngo, S, Innis, A, Hashem, Y, Boel, G. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Regulation of the macrolide resistance ABC-F translation factor MsrD.
Nat Commun, 14, 2023
|
|
3FE6
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand at pH 5.5 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
5HQD
 
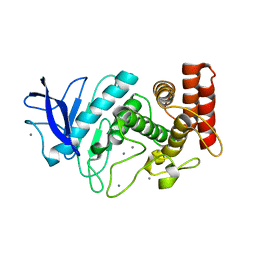 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
8GDW
 
 | |
7QP6
 
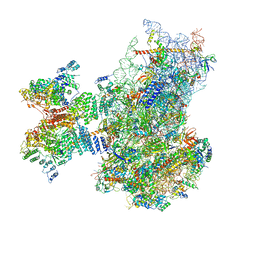 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
8GF4
 
 | |
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
6YAL
 
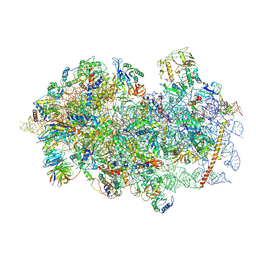 | | Mammalian 48S late-stage initiation complex with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S Ribosomal protein uS3, 40S Ribosomal protein uS7, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
