8DTB
 
 | |
8DRP
 
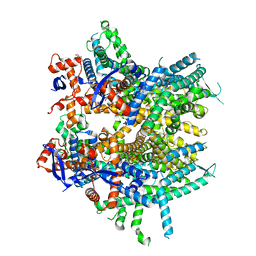 | |
8DJ1
 
 | | Crystal structure of NavAb V126T as a basis for the human Nav1.7 Inherited Erythromelalgia S241T mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for severe pain caused by mutations in the S4-S5 linkers of voltage-gated sodium channel Na V 1.7.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
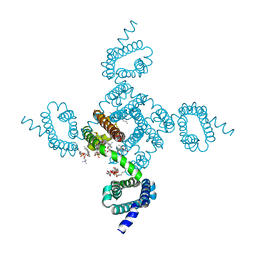
8DJ0
 
 | | Crystal structure of NavAb L123T as a basis for the human Nav1.7 Inherited Erythromelalgia I848T mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for severe pain caused by mutations in the S4-S5 linkers of voltage-gated sodium channel Na V 1.7.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
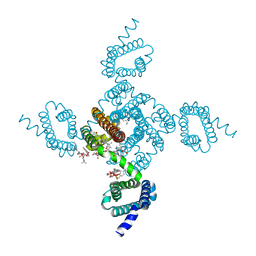
8DIZ
 
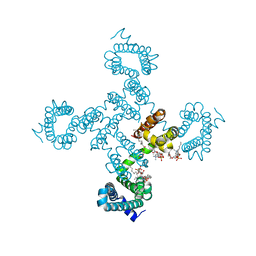 | | Crystal structure of NavAb I119T as a basis for the human Nav1.7 Inherited Erythromelalgia I234T mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for severe pain caused by mutations in the S4-S5 linkers of voltage-gated sodium channel Na V 1.7.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DIY
 
 | | Crystal structure of NavAb L101S as a basis for the human Nav1.7 Inherited Erythromelalgia F216S mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIX
 
 | | Structure of NavAb L98R as a basis for the human Nav1.7 Inherited Erythromelalgia L823R mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIW
 
 | | Crystal structure of NavAb E96P as a basis for the human Nav1.7 Inherited Erythromelalgia S211P mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIV
 
 | | Crystal structure of NavAb I22V as a basis for the human Nav1.7 Inherited Erythromelalgia I136V mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein, ... | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Powell, N.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DFL
 
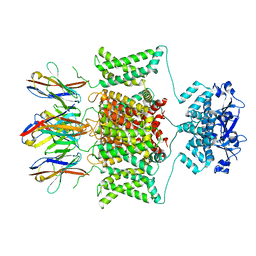 | |
8DDX
 
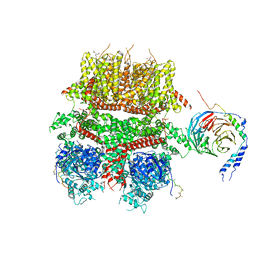 | | cryo-EM structure of TRPM3 ion channel in complex with Gbg in the presence of PIP2, tethered by ALFA-nanobody | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDW
 
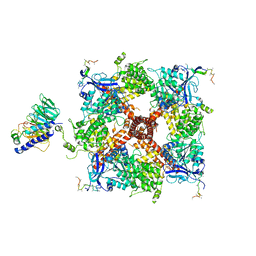 | |
8DDV
 
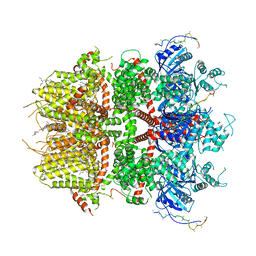 | | Cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDU
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDT
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDS
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDR
 
 | | cryo-EM structure of TRPM3 ion channel in the absence of PIP2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
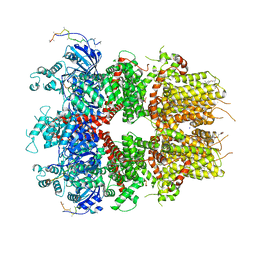 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8BX7
 
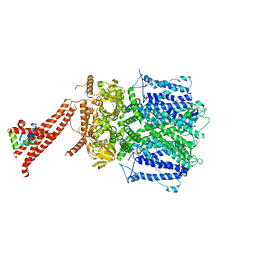 | | Structure of the rod CNG channel bound to calmodulin | | Descriptor: | Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1, calmodulin-1 | | Authors: | Marino, J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of calmodulin modulation of the rod cyclic nucleotide-gated channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BDC
 
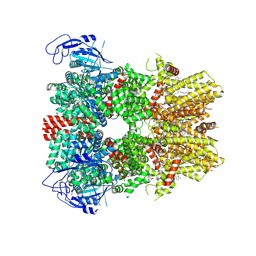 | | Human apo TRPM8 in a closed state (composite map) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Palchevskyi, S, Czarnocki-Cieciura, M, Vistoli, G, Gervasoni, S, Nowak, E, Beccari, A.R, Nowotny, M, Talarico, C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of human TRPM8 channel.
Commun Biol, 6, 2023
|
|
7ZJI
 
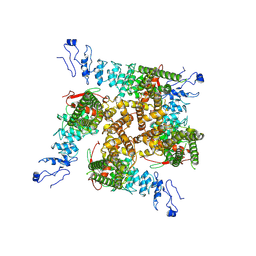 | |
7ZJH
 
 | | TRPV2-C16+Pro-2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJG
 
 | | Probenecid | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJE
 
 | | C16-2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2,Enhanced green fluorescent protein | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-10 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJD
 
 | |
