1GWN
 
 | | The crystal structure of the core domain of RhoE/Rnd3 - a constitutively activated small G protein | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho-related GTP-binding protein RhoE | | Authors: | Garavini, H, Riento, K, Phelan, J.P, McAlister, M.S.B, Ridley, A.J, Keep, N.H. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Core Domain of Rhoe/Rnd3: A Constitutively Activated Small G Protein
Biochemistry, 41, 2002
|
|
3J6U
 
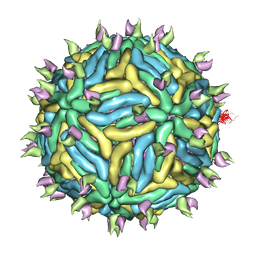 | | Cryo-EM structure of Dengue virus serotype 3 in complex with human antibody 5J7 Fab | | Descriptor: | Fab 5J7 heavy chain, Fab 5J7 light chain, envelope protein, ... | | Authors: | Fibriansah, G, Tan, J.L, Smith, S.A, de Alwis, R, Ng, T.-S, Kostyuchenko, V.A, Kukkaro, P, de Silva, A.M, Crowe Jr, J.E, Lok, S.-M. | | Deposit date: | 2014-03-26 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins.
Nat Commun, 6, 2015
|
|
3JW1
 
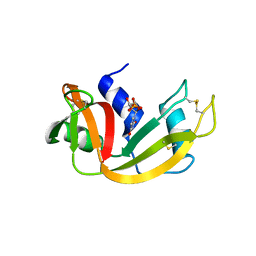 | | Crystal Structure of Bovine Pancreatic Ribonuclease Complexed with Uridine-5'-monophosphate at 1.60 A Resolution | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-MONOPHOSPHATE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic ribonuclease complexed with uridine 5'-monophosphate at 1.60 A resolution.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1H4V
 
 | |
3KB7
 
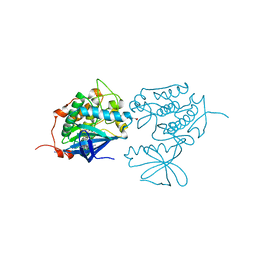 | | Crystal structure of Polo-like kinase 1 in complex with a pyrazoloquinazoline inhibitor | | Descriptor: | 8-{[2-methoxy-5-(4-methylpiperazin-1-yl)phenyl]amino}-1-methyl-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxamide, L(+)-TARTARIC ACID, Serine/threonine-protein kinase PLK1, ... | | Authors: | Bossi, R.T, Bertrand, J.A. | | Deposit date: | 2009-10-20 | | Release date: | 2010-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline derivatives as a new class of orally and selective Polo-like kinase 1 inhibitors
J.Med.Chem., 53, 2010
|
|
1HQ8
 
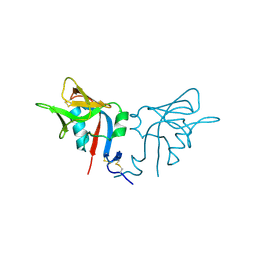 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
4V8Q
 
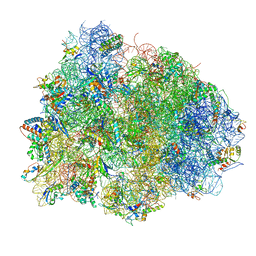 | | Complex of SmpB, a tmRNA fragment and EF-Tu-GDP-Kirromycin with the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Neubauer, C, Gillet, R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding in the absence of a codon by tmRNA and SmpB in the ribosome.
Science, 335, 2012
|
|
4V4I
 
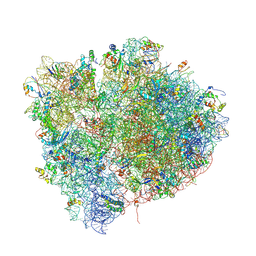 | | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements. | | Descriptor: | 16S SMALL SUBUNIT RIBOSOMAL RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-02-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements
Cell(Cambridge,Mass.), 126, 2006
|
|
6ZOZ
 
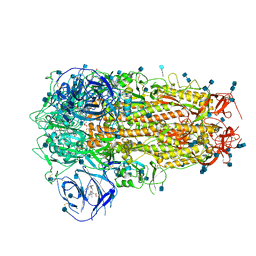 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3KZL
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
1QJT
 
 | | SOLUTION STRUCTURE OF THE APO EH1 DOMAIN OF MOUSE EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE 15, EPS15 | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE SUBSTRATE 15, EPS15 | | Authors: | Whitehead, B, Tessari, M, Carotenuto, A, van Bergen en Henegouwen, P.M, Vuister, G.W. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eh1 Domain of Eps15 is Structurally Classified as a Member of the S100 Subclass of EF-Hand Containing Proteins
Biochemistry, 38, 1999
|
|
3ZMF
 
 | | Salmonella enterica SadA 303-358 fused to GCN4 adaptors (SadAK2) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Albrecht, R, Lupas, A.N. | | Deposit date: | 2013-02-08 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A New Expression System for Protein Crystallization Using Trimeric Coiled-Coil Adaptors.
Protein Eng.Des.Sel., 21, 2008
|
|
1QRP
 
 | | Human pepsin 3A in complex with a phosphonate inhibitor IVA-VAL-VAL-LEU(P)-(O)PHE-ALA-ALA-OME | | Descriptor: | PEPSIN 3A, methyl N-[(2S)-2-({(S)-hydroxy[(1R)-3-methyl-1-{[N-(3-methylbutanoyl)-L-valyl-L-valyl]amino}butyl]phosphoryl}oxy)-3-phenylpropanoyl]-L-alanyl-L-alaninate | | Authors: | Fujinaga, M, Cherney, M.M, Tarasova, N.I, Bartlett, P.A, Hanson, J.E, James, M.N.G. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2PXQ
 
 | | Variant 14 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
6ZJF
 
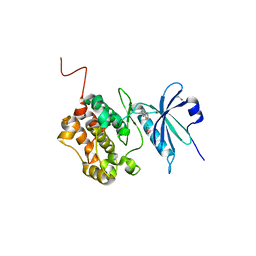 | | Crystal structure of STK17B (DRAK2) in complex with AP-229 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-cyclopropylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Picado, A, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
4V5C
 
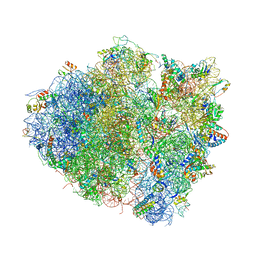 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
2AA2
 
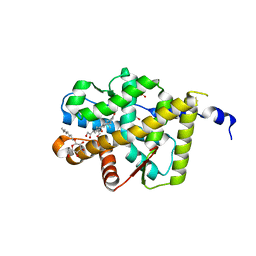 | | Mineralocorticoid Receptor with Bound Aldosterone | | Descriptor: | ALDOSTERONE, GLYCEROL, Mineralocorticoid receptor, ... | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
6YUU
 
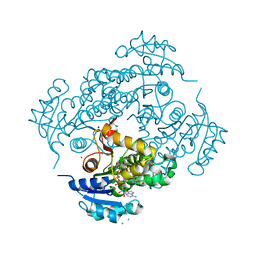 | | Crystal structure of M. tuberculosis InhA inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Weinrich, J.D, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
6Z6C
 
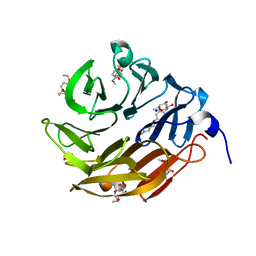 | | Crystal structurel of FleA lectin in complex with a monovalent inhibitor | | Descriptor: | 4-((1-(2-(2-(2-(2-hydroxyethoxy)ethoxy)ethoxy)ethyl)-1H-1,2,3-triazol-4-yl)methoxy)benzyl-a-L-thiofucoside, Fucose-specific lectin, GLYCEROL, ... | | Authors: | Varrot, A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hexavalent thiofucosides to probe the role of the Aspergillus fumigatus lectin FleA in fungal pathogenicity.
Org.Biomol.Chem., 19, 2021
|
|
4V5D
 
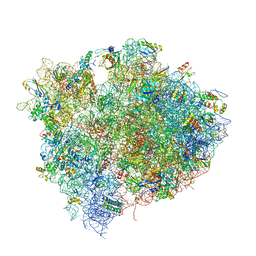 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
7QRM
 
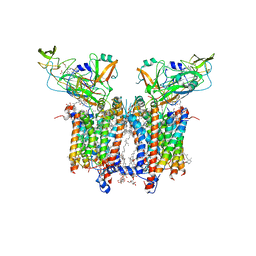 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.7 A resolution | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Sarewicz, M, Szwalec, M, Indyka, P, Rawski, M, Pintscher, S, Pietras, R, Mielecki, B, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
3KFK
 
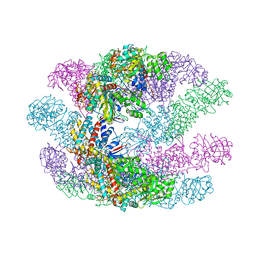 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (6.003 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3ZMK
 
 | |
1HQ1
 
 | | STRUCTURAL AND ENERGETIC ANALYSIS OF RNA RECOGNITION BY A UNIVERSALLY CONSERVED PROTEIN FROM THE SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Sagar, M.B, Doudna, J.A. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle.
J.Mol.Biol., 307, 2001
|
|
3KHW
 
 | |
