5NBO
 
 | | Bacteroides ovatus mixed linkage glucan PUL (MLGUL) GH16 | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 16 | | Authors: | Hemsworth, G.R, Tamura, K, Dejean, G, Rogers, T.E, Pudlo, N.A, Urs, K, Jain, N, Martens, E.C, Brumer, H, Davies, G.J. | | Deposit date: | 2017-03-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides.
Cell Rep, 21, 2017
|
|
6S7B
 
 | | Crystal structure of CARM1 in complex with inhibitor UM249 | | Descriptor: | 1-[4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]butyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
5NCK
 
 | |
6S7Q
 
 | |
6RV8
 
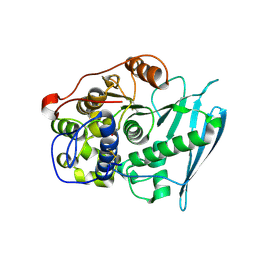 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
5NK8
 
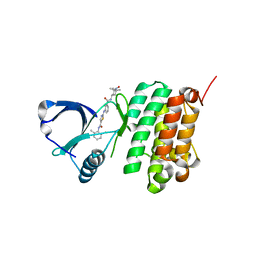 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2f | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[(2,2,6,6-tetramethyl-1-oxidanyl-piperidin-4-yl)carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NE1
 
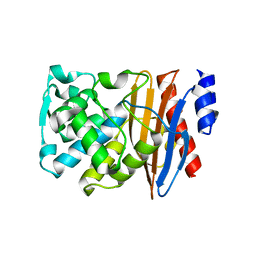 | | L2 class A serine-beta-lactamase in complex with cyclic boronate 2 | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Beta-lactamase, TRIETHYLENE GLYCOL | | Authors: | Hinchliffe, P, Calvopina, K, Spencer, J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates.
Mol. Microbiol., 106, 2017
|
|
6RVU
 
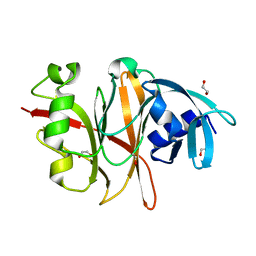 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
5NKG
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3d | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-ethyl-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKP
 
 | | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide | | Descriptor: | CHLORIDE ION, CITRIC ACID, Kelch-like protein 3, ... | | Authors: | Chen, Z, Sorrell, F.J, Pinkas, D.M, Williams, E, Mathea, S, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Burgess-Brown, N, Bountra, C, Bullock, A. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide
To Be Published
|
|
6RKB
 
 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 1 | | Descriptor: | 4-[(~{E})-2-(4-fluorophenyl)ethenyl]-1-[(~{E})-prop-1-enyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
6RKP
 
 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 84 | | Descriptor: | 1-[(~{E})-prop-1-enyl]-4-[(~{E})-2-[4-(trifluoromethyl)phenyl]ethenyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
6RXA
 
 | | EDDS lyase variant D290M/Y320M with bound formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, GLYCEROL, ... | | Authors: | Grandi, E, Poelarends, G.J, Thunnissen, A.M.W.H. | | Deposit date: | 2019-06-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Engineered C-N Lyase: Enantioselective Synthesis of Chiral Synthons for Artificial Dipeptide Sweeteners.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6S1H
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
5NMC
 
 | |
6S29
 
 | |
5NMX
 
 | | Crystal Structure of the pyrrolizidine alkaloid N-oxygenase from Zonocerus variegatus in complex with FAD and NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, MAGNESIUM ION, ... | | Authors: | Scheidig, A, Kubitza, C, Faust, A, Ober, D. | | Deposit date: | 2017-04-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6S2Q
 
 | |
6RMM
 
 | |
5NNR
 
 | | Structure of Naa15/Naa10 bound to HypK-THB | | Descriptor: | HypK, N-terminal acetyltransferase-like protein, Naa10 | | Authors: | Weyer, F.A, Gumiero, A, Kopp, J, Sinning, I. | | Deposit date: | 2017-04-10 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of HypK regulating N-terminal acetylation by the NatA complex.
Nat Commun, 8, 2017
|
|
5NOH
 
 | | HRSV M2-1 core domain | | Descriptor: | Transcription elongation factor M2-1 | | Authors: | Josts, I, Almeida Hernandez, Y, Molina, I.G, de Prat-Gay, G, Tidow, H. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5NON
 
 | | Structure of truncated Norcoclaurine Synthase from Thalictrum flavum with product mimic | | Descriptor: | 4-[2-[2-(4-methoxyphenyl)ethylamino]ethyl]benzene-1,2-diol, S-norcoclaurine synthase | | Authors: | Sula, A, Lichman, B.R, Pesnot, T, Ward, J.M, Hailes, H.C, Keep, N.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Evidence for the Dopamine-First Mechanism of Norcoclaurine Synthase.
Biochemistry, 56, 2017
|
|
6S3I
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-MgF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
6S49
 
 | |
5NG2
 
 | | Structure of RIP2K(D146N) with bound Staurosporine | | Descriptor: | PHOSPHATE ION, Receptor-interacting serine/threonine-protein kinase 2, STAUROSPORINE | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
