1XNN
 
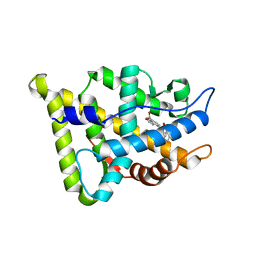 | | CRYSTAL STRUCTURE OF THE RAT ANDROGEN RECEPTOR LIGAND BINDING DOMAIN T877A MUTANT COMPLEX WITH (3A-ALPHA-,4-ALPHA 7-ALPHA-,7A-ALPHA-)-3A,4,7,7A-TETRAHYDRO-2-(4-NITRO-1-NAPHTHALENYL)-4,7-ETHANO-1H-ISOINDOLE-1,3(2H)-DIONE. | | Descriptor: | Androgen receptor, REL-(3AR,4S,7R,7AS)-3A,4,7,7A-TETRAHYDRO-2-(4-NITRO-1-NAPHTHALENYL)-4,7-ETHANO-1H-ISOINDOLE-1,3(2H)-DIONE | | Authors: | Sack, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based approach to the design of bicyclic-1H-isoindole-1,3(2H)-dione based androgen receptor antagonists.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1BZE
 
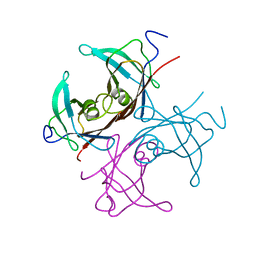 | |
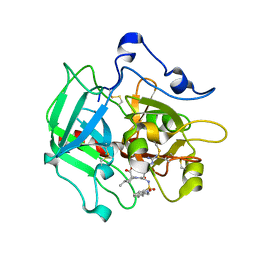
3R6B
 
 | | Crystal Structure of Thrombospondin-1 TSR Domains 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1 | | Authors: | Page, R.C, Klenotic, P.A, Misra, S, Silverstein, R.L. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system.
Protein Expr.Purif., 80, 2011
|
|
4RN6
 
 | | Structure of prethrombin-2 mutant s195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Thrombin heavy chain | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Niu, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
2WD7
 
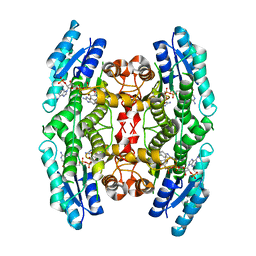 | | PTERIDINE REDUCTASE 1 (PTR1) FROM TRYPANOSOMA BRUCEI IN COMPLEX WITH NADP AND DDD00066750 | | Descriptor: | 6-chloro-1H-benzimidazol-2-amine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Robinson, D.A, Wyatt, P.G, Spinks, D, Brenk, R. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | One Scaffold, Three Binding Modes: Novel and Selective Pteridine Reductase 1 Inhibitors Derived from Fragment Hits Discovered by Virtual Screening.
J.Med.Chem., 52, 2009
|
|
6KBF
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 3 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydro-2~{H}-isoquinolin-1-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
4BKE
 
 | | Recombinant human serum albumin with palmitic acid. Synthetic cationic antimicrobial peptides bind with their hydrophobic parts to drug site II of human serum albumin | | Descriptor: | PALMITIC ACID, SERUM ALBUMIN | | Authors: | Sivertsen, A, Isaksson, J, Leiros, H.-K.S, Svenson, J, Svendsen, J.-S, Brandsdal, B.-O. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthetic Cationic Antimicrobial Peptides Bind with Their Hydrophobic Parts to Drug Site II of Human Serum Albumin.
Bmc Struct.Biol., 14, 2014
|
|
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
4Q7W
 
 | |
4O77
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB 202190 | | Descriptor: | 4-[4-(4-fluorophenyl)-5-(pyridin-4-yl)-1H-imidazol-2-yl]phenol, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
3U8W
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Triazolopyridazinone inhibitor | | Descriptor: | 3-[3-(2-chloro-6-fluorophenyl)-5-ethyl-6-oxo-5,6-dihydro[1,2,4]triazolo[4,3-b]pyridazin-7-yl]-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of triazolopyridazinones as potent p38alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2Y9M
 
 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
1Y5V
 
 | | tRNA-Guanine Transglycosylase (TGT) in complex with 6-Amino-4-(2-phenylethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-(2-PHENYLETHYL)-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
6KO9
 
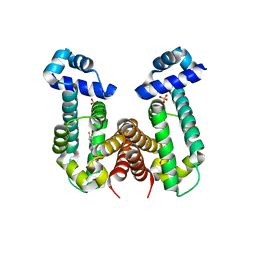 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
1OK8
 
 | |
6KJV
 
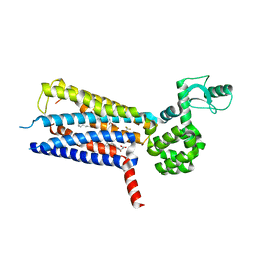 | | Structure of thermal-stabilised(M9) human GLP-1 receptor transmembrane domain | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
6KO8
 
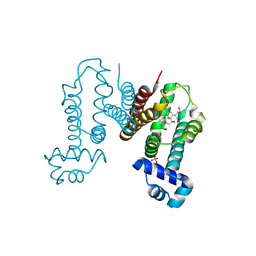 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
3ONH
 
 | | Crystal structure of UBA2ufd-Ubc9: insights into E1-E2 interactions in Sumo pathways | | Descriptor: | Ubiquitin-activating enzyme E1-like | | Authors: | Wang, J, Taherbhoy, A.M, Hunt, H.W, Seyedin, S.N, Miller, D.W, Huang, D.T, Schulman, B.A. | | Deposit date: | 2010-08-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.
Plos One, 5, 2010
|
|
2LAS
 
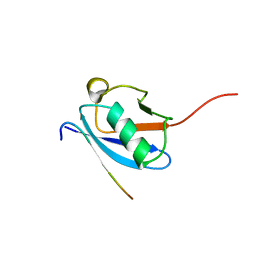 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
4QOQ
 
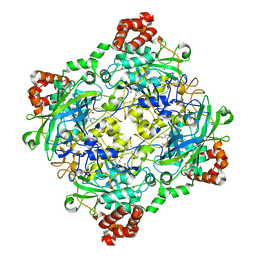 | | Structure of Bacillus pumilus catalase with guaiacol bound | | Descriptor: | CHLORIDE ION, Catalase, Guaiacol, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | Descriptor: | CATECHOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
1Y6R
 
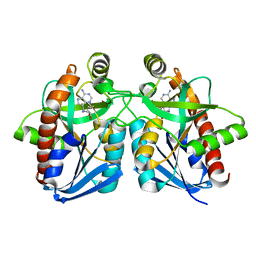 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
3ASL
 
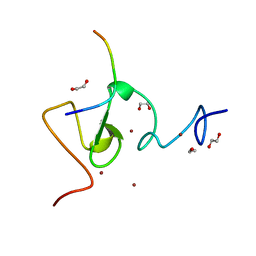 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6L15
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
