3IDP
 
 | | B-Raf V600E kinase domain in complex with an aminoisoquinoline inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N~1~-(4-chlorophenyl)-6-methyl-N~5~-[3-(7H-purin-6-yl)pyridin-2-yl]isoquinoline-1,5-diamine | | Authors: | Whittington, D.A, Epstein, L.F. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective inhibitors of the mutant B-raf pathway: discovery of a potent and orally bioavailable aminoisoquinoline.
J.Med.Chem., 52, 2009
|
|
5LA4
 
 | | Crystal structure of apo human proheparanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heparanase, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5UJW
 
 | | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, CITRIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
4FR4
 
 | | Crystal structure of human serine/threonine-protein kinase 32A (YANK1) | | Descriptor: | 1,2-ETHANEDIOL, STAUROSPORINE, Serine/threonine-protein kinase 32A | | Authors: | Chaikuad, A, Elkins, J.M, Krojer, T, Mahajan, P, Goubin, S, Szklarz, M, Tumber, A, Wang, J, Savitsky, P, Shrestha, B, Daga, N, Picaud, S, Fedorov, O, Allerston, C.K, Latwiel, S.V.A, Vollmar, M, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human serine/threonine-protein kinase 32A (YANK1)
To be Published
|
|
2ON7
 
 | | Structure of NaGST-1 | | Descriptor: | Na Glutathione S-transferase 1 | | Authors: | Asojo, O.A, Ngamelue, M, Homma, H, Goud, G, Zhan, B, Hotez, P.J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of Na-GST-1 and Na-GST-2 two glutathione s-transferase from the human hookworm Necator americanus
Bmc Struct.Biol., 7, 2007
|
|
6F7I
 
 | | human MALT1(329-728) IN COMPLEX WITH MLT-747 | | Descriptor: | 1-[2-chloranyl-7-[(1~{S})-1-methoxyethyl]pyrazolo[1,5-a]pyrimidin-6-yl]-3-(5-chloranyl-6-pyrrolidin-1-ylcarbonyl-pyridin-3-yl)urea, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Renatus, M, Renatus, M. | | Deposit date: | 2017-12-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient.
Nat. Chem. Biol., 15, 2019
|
|
5UUZ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P200 | | Descriptor: | 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P200
To Be Published
|
|
5UZV
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with 5' recessed-end DNA (rI) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V0C
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with 5' flap DNA (f2I) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*A)-3'), DNA (5'-D(P*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3E0Q
 
 | | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase complexed with a novel monocyclic inhibitor | | Descriptor: | 6-amino-5-bromo-1,2,3,4-tetrahydropyrimidine-2,4-dione, DIMETHYL SULFOXIDE, Purine-nucleoside phosphorylase | | Authors: | Pereira, H.M, Berdini, V, Ferri, M.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2008-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Schistosoma purine nucleoside phosphorylase complexed with a novel monocyclic inhibitor.
Acta Trop., 114, 2010
|
|
1LIB
 
 | |
5Y8Y
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
8XBC
 
 | | The substrate binding protein of an ABC transporter in complex with beta-1,3-xylotriose | | Descriptor: | XylA, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-beta-D-xylopyranose | | Authors: | Zhao, F, Sun, H.N, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2023-12-06 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Complete xylan utilization pathway and regulation mechanisms involved in marine algae degradation by cosmopolitan marine and human gut microbiota.
Isme J, 19, 2025
|
|
2AI6
 
 | |
4BGH
 
 | | Crystal Structure of CDK2 in complex with pan-CDK Inhibitor | | Descriptor: | 4-((5-BROMO-4-(PROP-2-YN-1-YLAMINO)PYRIMIDIN-2-YL)AMINO)BENZENESULFONAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Luecking, U, Jautelat, R, Krueger, M, Brumby, T, Lienau, P, Schaefer, M, Briem, H, Schulze, J, Hillisch, A, Reichel, A, Siemeister, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Lab Oddity Prevails: Discovery of Pan-Cdk Inhibitor (R)- S-Cyclopropyl-S-(4-{[4-{[(1R,2R)-2-Hydroxy-1-Methylpropyl]Oxy}-5-(Trifluoromethyl)Pyrimidin-2-Yl]Amino}Phenyl)Sulfoximide (Bay 1000394) for the Treatment of Cancer.
Chemmedchem, 8, 2013
|
|
3AOK
 
 | |
3AOX
 
 | | X-ray crystal structure of human anaplastic lymphoma kinase in complex with CH5424802 | | Descriptor: | 1,2-ETHANEDIOL, 9-ethyl-6,6-dimethyl-8-[4-(morpholin-4-yl)piperidin-1-yl]-11-oxo-6,11-dihydro-5H-benzo[b]carbazole-3-carbonitrile, ALK tyrosine kinase receptor | | Authors: | Nagel, S, Moertl, M, Jestel, A, Fukami, T.A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant
Cancer Cell, 19, 2011
|
|
4KV2
 
 | |
7UPL
 
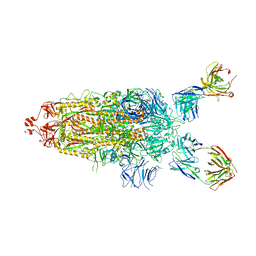 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7C6K
 
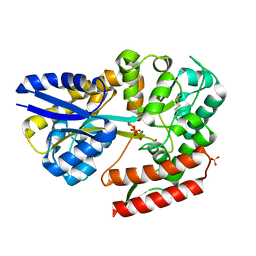 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotriose (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PYROPHOSPHATE 2-, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
9GHG
 
 | |
3IXQ
 
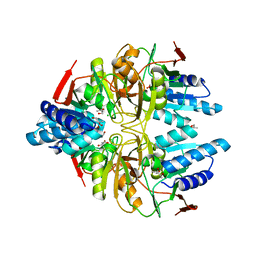 | | Structure of ribose 5-phosphate isomerase a from methanocaldococcus jannaschii | | Descriptor: | ACETATE ION, CHLORIDE ION, Ribose-5-phosphate isomerase A, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of an archaeal ribose-5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
9GHH
 
 | |
7C6L
 
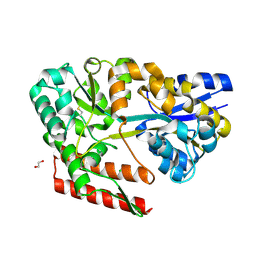 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotriose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5V0B
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with 5' recessed-end DNA (rIX) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
