7BB2
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
2Q2I
 
 | | Crystal structure of the protein secretion chaperone CsaA from Agrobacterium tumefaciens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Secretion chaperone | | Authors: | Feldman, A.R, Shapova, Y.A, Paetzel, M. | | Deposit date: | 2007-05-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.
J.Mol.Biol., 379, 2008
|
|
2AFZ
 
 | | Crystal structure of human glutaminyl cyclase in complex with 1-vinylimidazole | | Descriptor: | 1-VINYLIMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
6EVE
 
 | | Structure of R175A S. cerevisiae Fdc1 with prFMN in the iminium form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
3IT4
 
 | | The Crystal Structure of Ornithine Acetyltransferase from Mycobacterium tuberculosis (Rv1653) at 1.7 A | | Descriptor: | ACETATE ION, Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, ... | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
6IBL
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
3IUE
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(5-methoxy-2-(5-Methylpyridin-2-ylsulfonylcarbamoyl)-1H-indol-1-yl) acetic acid | | Descriptor: | (5-methoxy-2-{[(5-methylpyridin-2-yl)sulfonyl]carbamoyl}-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Wen, S, Hung, A.W, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6QR5
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-[1-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazol-5-amine, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
1IWD
 
 | |
1IYL
 
 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
6IDW
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2018-09-11 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structures of the GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo and endo activity and its complex with cellobiose.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1WVN
 
 | |
7XRL
 
 | | Diol dehydratase complexed with AdoMeCbl and 1,2-propanediol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XUC
 
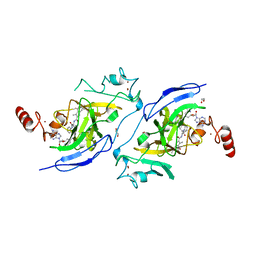 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
6QMK
 
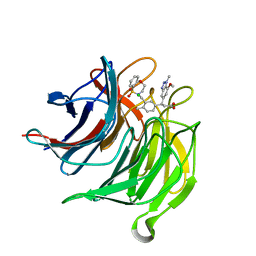 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-[3-[[1,1-bis(oxidanylidene)-3,4-dihydro-5,1$l^{6},2-benzoxathiazepin-2-yl]methyl]-4-methyl-phenyl]-3-(7-methoxy-1-methyl-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
9EY5
 
 | |
6B1K
 
 | | Macrophage Migration Inhibitory Factor in Complex with a Naphthyridinone Inhibitor (3a) | | Descriptor: | 2-[1-(4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]-7-methyl-1,7-naphthyridin-8(7H)-one, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Krimmer, S.G, Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Adding a Hydrogen Bond May Not Help: Naphthyridinone vs Quinoline Inhibitors of Macrophage Migration Inhibitory Factor.
ACS Med Chem Lett, 8, 2017
|
|
5N7J
 
 | |
3IVC
 
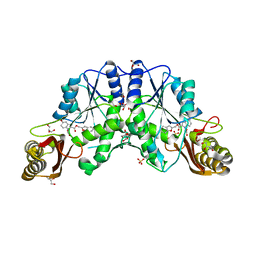 | | Crystal structure of pantothenate synthetase in complex with 2-(2-((benzofuran-2-ylmethoxy)carbonyl)-5-methoxy-1H-indol-1-yl)acetic acid | | Descriptor: | ETHANOL, GLYCEROL, Pantothenate synthetase, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6QR6
 
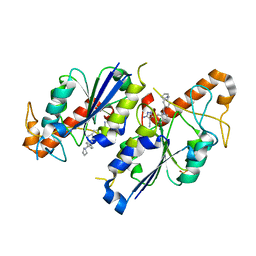 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
4ZT7
 
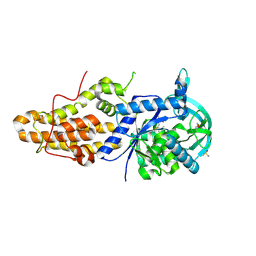 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor N-[(4R)-6,8-dichloro-1,2,3,4-tetrahydroquinolin-4-yl]-N'-(5-fluoro-3H-imidazo[4,5-b]pyridin-2-yl)propane-1,3-diamine (Chem 1717) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, METHIONINE, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-05-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
Acs Infect Dis., 2, 2016
|
|
5U3B
 
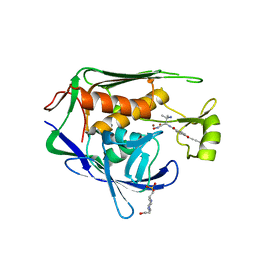 | | Pseudomonas aeruginosa LpxC in complex with NVS-LPXC-01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[(but-2-yn-1-yl)oxy]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
5G4S
 
 | | BROMODOMAIN OF HUMAN BRPF1 WITH N-1,3-dimethyl-6-2R-2- methylpiperazin-1-yl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl-N- ethyl-2-methoxybenzamide | | Descriptor: | 1,2-ETHANEDIOL, N-[1,3-dimethyl-6-[(2R)-2-methylpiperazin-1-yl]-2-oxidanylidene-benzimidazol-5-yl]-N-ethyl-2-methoxy-benzamide, PEREGRIN | | Authors: | Chung, C. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gsk6853, a Chemical Probe for Inhibition of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 7, 2016
|
|
8DU5
 
 | | Murine sialidase-1 (NEU1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialidase-1, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2022-07-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the immunoregulatory sialidase NEU1.
Sci Adv, 9, 2023
|
|
