2NGR
 
 | | TRANSITION STATE COMPLEX FOR GTP HYDROLYSIS BY CDC42: COMPARISONS OF THE HIGH RESOLUTION STRUCTURES FOR CDC42 BOUND TO THE ACTIVE AND CATALYTICALLY COMPROMISED FORMS OF THE CDC42-GAP. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G, Clardy, J, Cerione, R. | | Deposit date: | 1998-07-31 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
1WRY
 
 | | Solution structure of the SH3 domain-binding glutamic acid-rich-like protein | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Inoue, K, Miyamoto, K, Nagashima, T, Hayashi, F, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-29 | | Release date: | 2005-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain-binding glutamic acid-rich-like protein
To be Published
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
3VHL
 
 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
1WGM
 
 | | Solution structure of the U-box in human ubiquitin conjugation factor E4A | | Descriptor: | Ubiquitin conjugation factor E4A | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U-box in human ubiquitin conjugation factor E4A
To be Published
|
|
1AN0
 
 | | CDC42HS-GDP COMPLEX | | Descriptor: | CDC42HS-GDP, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kongsaeree, P, Cerione, R, Clardy, J. | | Deposit date: | 1997-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure Determination of Cdc42Hs and Gdp Complex
To be Published
|
|
7OLD
 
 | | Thermophilic eukaryotic 80S ribosome at pe/E (TI)-POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
7OLC
 
 | | Thermophilic eukaryotic 80S ribosome at idle POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
1CF4
 
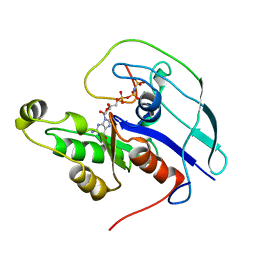 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
7PWG
 
 | | Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L18a, 60S ribosomal protein L27, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
7PWO
 
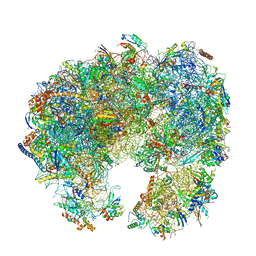 | | Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S26, 40S ribosomal protein S30, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
3ITE
 
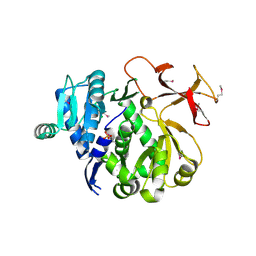 | | The third adenylation domain of the fungal SidN non-ribosomal peptide synthetase | | Descriptor: | CHLORIDE ION, SULFATE ION, SidN siderophore synthetase | | Authors: | Lee, T.V, Lott, J.S, Johnson, R.D, Johnson, L.J, Arcus, V.L. | | Deposit date: | 2009-08-28 | | Release date: | 2009-11-17 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
J.Biol.Chem., 285, 2010
|
|
1CEE
 
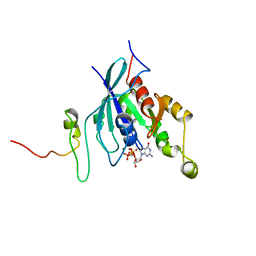 | | SOLUTION STRUCTURE OF CDC42 IN COMPLEX WITH THE GTPASE BINDING DOMAIN OF WASP | | Descriptor: | GTP-BINDING RHO-LIKE PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Abdul-Manan, N, Aghazadeh, B, Liu, G.A, Majumdar, A, Ouerfelli, O, Rosen, M.K. | | Deposit date: | 1999-03-08 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Nature, 399, 1999
|
|
1E0A
 
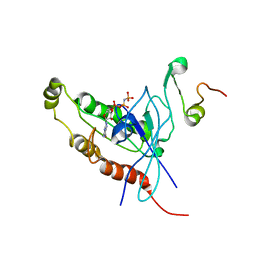 | | Cdc42 complexed with the GTPase binding domain of p21 activated kinase | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Morreale, A, Venkatesan, M, Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Laue, E.D. | | Deposit date: | 2000-03-16 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cdc42 Bound to the Gtpase Binding Domian of Pak
Nat.Struct.Biol., 7, 2000
|
|
1DOA
 
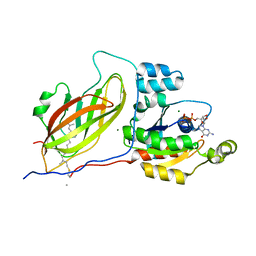 | | Structure of the rho family gtp-binding protein cdc42 in complex with the multifunctional regulator rhogdi | | Descriptor: | GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hoffman, G.R, Nassar, N, Cerione, R.C. | | Deposit date: | 1999-12-20 | | Release date: | 2000-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI.
Cell(Cambridge,Mass.), 100, 2000
|
|
1EES
 
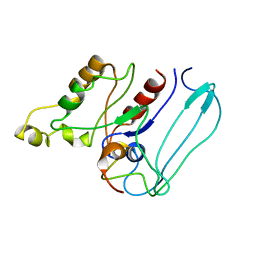 | | SOLUTION STRUCTURE OF CDC42HS COMPLEXED WITH A PEPTIDE DERIVED FROM P-21 ACTIVATED KINASE, NMR, 20 STRUCTURES | | Descriptor: | GTP-BINDING PROTEIN, P21-ACTIVATED KINASE | | Authors: | Gizachew, D, Guo, W, Chohan, K.C, Sutcliffe, M.J, Oswald, R.E. | | Deposit date: | 2000-02-02 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase.
Biochemistry, 39, 2000
|
|
1A4R
 
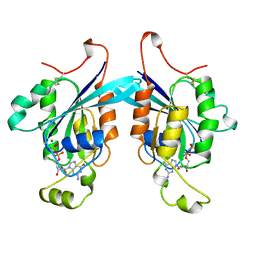 | | G12V MUTANT OF HUMAN PLACENTAL CDC42 GTPASE IN THE GDP FORM | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, G25K GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rudolph, M.G, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 1998-02-02 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleotide binding to the G12V-mutant of Cdc42 investigated by X-ray diffraction and fluorescence spectroscopy: two different nucleotide states in one crystal.
Protein Sci., 8, 1999
|
|
3A4R
 
 | | The crystal structure of SUMO-like domain 2 in Nip45 | | Descriptor: | 1,2-ETHANEDIOL, NFATC2-interacting protein, SULFATE ION | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
6XIQ
 
 | | Cryo-EM Structure of K63R Ubiquitin Mutant Ribosome under Oxidative Stress | | Descriptor: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhou, Y, Bartesaghi, A, Silva, G.M. | | Deposit date: | 2020-06-21 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1U0G
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0F
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0E
 
 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
6N8M
 
 | | Cryo-EM structure of pre-Lsg1 (PL) pre-60S ribosomal subunit | | Descriptor: | 5.8S RNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8N
 
 | | Cryo-EM structure of Lsg1-engaged (LE) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
