7BYS
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
6T9S
 
 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1S)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | Descriptor: | (2~{E})-2-hydroxyimino-~{N}-[(1~{S})-3-[4-[(2-methylimidazol-1-yl)methyl]-1,2,3-triazol-1-yl]-1-phenyl-propyl]ethanamid e, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(5-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
2J5T
 
 | | Glutamate 5-kinase from Escherichia coli complexed with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE 5-KINASE, GLUTAMIC ACID, ... | | Authors: | Marco-Marin, C, Gil-Ortiz, F, Perez-Arellano, I, Cervera, J, Fita, I, Rubio, V. | | Deposit date: | 2006-09-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Two-Domain Architecture within the Amino Acid Kinase Enzyme Family Revealed by the Crystal Structure of Escherichia Coli Glutamate 5-Kinase.
J.Mol.Biol., 367, 2007
|
|
7BZM
 
 | | Crystal structure of rice Os3BGlu7 with glucoimidazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLUCOIMIDAZOLE, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R, Tankrathok, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7.
Biomolecules, 10, 2020
|
|
2J70
 
 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
6TAD
 
 | | Bd0314 DslA E143Q mutant | | Descriptor: | SLT domain-containing protein | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
7C09
 
 | |
6TC6
 
 | |
5X0R
 
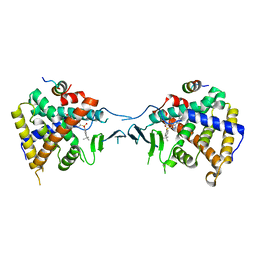 | | Crystal Structure of PXR LBD Complexed with SJB7 | | Descriptor: | 4-[(4-tert-butylphenyl)sulfonyl]-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Lv, L, Lin, W, Chai, S.C, Zhang, Q, Chen, T. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | SPA70 is a potent antagonist of human pregnane X receptor.
Nat Commun, 8, 2017
|
|
6TN1
 
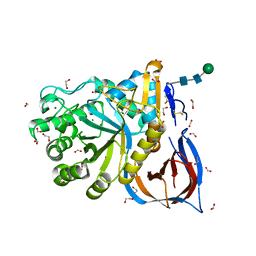 | | Unliganded Crystal Structure of Recombinant GBA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7BU6
 
 | | Structure of human beta1 adrenergic receptor bound to norepinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
6TEF
 
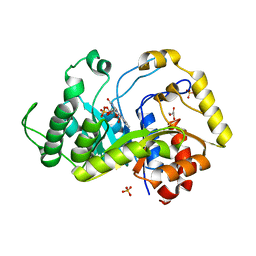 | | Crystal structure of monooxygenase RutA complexed with dioxygen under 0.5 MPa / 5 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-11-11 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
2J8U
 
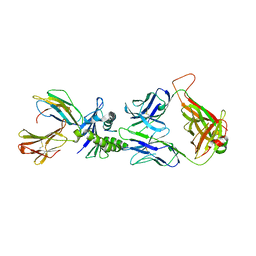 | | Large CDR3a loop alteration as a function of MHC mutation. | | Descriptor: | AHIII TCR ALPHA CHAIN, AHIII TCR BETA CHAIN, Beta-2-microglobulin, ... | | Authors: | Miller, P.J, Benhar, Y.P, Biddison, W, Collins, E.J. | | Deposit date: | 2006-10-27 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Single MHC mutation eliminates enthalpy associated with T cell receptor binding.
J. Mol. Biol., 373, 2007
|
|
2J6O
 
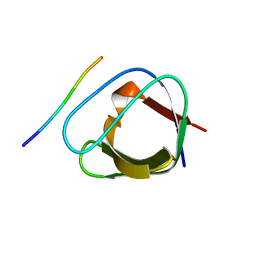 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETEROTRIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
6IJ2
 
 | |
2VUE
 
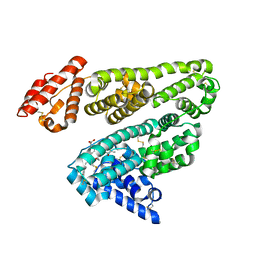 | | Human serum albumin complexed with 4Z,15E-bilirubin-IX-alpha | | Descriptor: | BILIVERDINE IX ALPHA, SERUM ALBUMIN | | Authors: | Zunszain, P.A, Ghuman, J, Curry, S. | | Deposit date: | 2008-05-24 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystallographic Analysis of Human Serum Albumin Complexed with 4Z,15E-Bilirubin-Ixalpha.
J.Mol.Biol., 381, 2008
|
|
2J6F
 
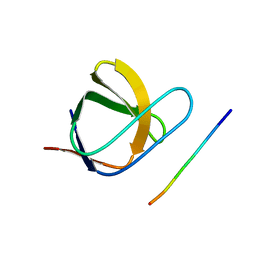 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) BOUND TO CBL-B PEPTIDE | | Descriptor: | CD2-ASSOCIATED PROTEIN, E3 UBIQUITIN-PROTEIN LIGASE CBL-B | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-09-28 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2N72
 
 | | Solution structure of the Q domain from ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | Veverka, V, Hexnerova, R. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Sci Rep, 6, 2016
|
|
6IQR
 
 | |
2MSU
 
 | |
2IVP
 
 | | Structure of UP1 protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
7BR3
 
 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
5Y6E
 
 | |
6HCQ
 
 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
